乳酸菌(lactic acid bacteria, LAB)是一类能发酵碳水化合物生成乳酸的细菌总称,主要包括乳杆菌属(Lactobacillus)、链球菌属(Streptococcus)、肠球菌属(Enterococcus)、乳球菌属(Lactococcus)、双歧杆菌属(Bifidobacterium)和明串珠菌属(Leuconostoc)[1]。乳酸菌能够减轻乳糖不耐症、减轻病毒和药物引起的腹泻、辅助治疗溃疡、调节免疫系统[1]、防治龋齿、预防II型糖尿病和肥胖以及降低血清胆固醇水平[2]。乳酸菌在食品工业中应用广泛,例如生产乳制品、肉制品、泡菜、饮料和天然防腐剂。酸奶发酵剂主要菌种是保加利亚乳杆菌(Lactobacillus delbrueckii subsp.bulgaricus)和嗜热链球菌(Streptococcus thermophilus),其他乳酸菌作为辅助,以不同比例共生形成用于酸奶发酵的微生物菌群[3]。
双组分信号转导系统(two-component signal transduction system, TCS)是细菌中重要的信号机制,经典TCS由跨膜受体组氨酸蛋白激酶(histidine protein kinase, HPK)和同源胞质内应答调控蛋白(response regulator protein, RR)组成,细菌通过TCS将环境刺激与适应性反应相结合,调控新陈代谢和运动等基本生理活动以及发育和毒力等特殊过程。HPK感应外界信号刺激后磷酸化自身保守的组氨酸残基,将磷酸化基团转移到RR的保守天冬氨酸残基上,从而激活RR效应结构域进行应答调控[4](图1)。目前已有许多有关TCS的报道,例如EnvZ/OmpR TCS调控大肠杆菌OmpC蛋白表达,在乙醇耐受中起关键作用[5];SalK/SalR、Ihk/Irr、VirR/VirS、NisK/NisR、1910HK/RR等TCS与猪链球菌毒力相关[6];PhoP/PhoQ TCS可以感知二价阳离子、低pH和阳离子抗菌肽的存在,调控大肠杆菌或沙门氏菌中Mg2+稳态、细胞膜组成、抗逆性和毒力[7]。

图1 经典TCS
Fig.1 Classical two-component signal transduction system
1 保加利亚乳杆菌TCS
作为最重要的工业乳酸菌,保加利亚乳杆菌能利用牛奶中乳糖厌氧发酵快速合成乳酸,因此在发酵乳制品生产中被广泛使用。不同保加利亚乳杆菌的TCS数量有所差异,如保加利亚乳杆菌ATCC BAA 365具有7对完整的TCS,而ATCC 11842菌株仅有5对完整的TCS及1个孤立HPK和1个孤立RR[8-9]。
1.1 调控酸适应性
在酸奶发酵过程中生长环境明显酸化,pH值从6.5下降到4.2左右。因此,酸适应能力和耐受性对保加利亚乳杆菌具有重要意义[10]。CUI等[11]采用定量聚合酶链反应(quantitative PCR, qPCR)方法评估保加利亚乳杆菌CH3菌株TCS在酸适应过程中的基因表达变化,发现2个TCS JN675228/JN675229和JN675230/JN675231及2个HPK编码基因JN675236和JN675240表达量上调,推测其与酸适应能力有关。为验证此推测,作者选取TCS JN675228(HPK编码基因)/JN675229(RR编码基因)进行敲除,2个突变菌株的生长能力均比野生型显著下降;JN675228敲除对菌株酸适应能力影响不大,而JN675229敲除使菌株酸适应能力下降。酵母双杂交实验也证实JN675228和JN675229编码蛋白之间存在直接相互作用。这些结果表明保加利亚乳杆菌中TCS JN675228/JN675229与酸适应能力密切相关。为进一步研究JN675228/JN675229在保加利亚乳杆菌酸适应的信号转导机制,WANG等[10]采用二维凝胶电泳和飞行时间质谱法,筛选出在自然发酵和酸适应条件下JN675228/JN675229基因敲除突变株与野生菌株的差异表达蛋白,并结合转录数据鉴定JN675228/JN675229的靶基因(表1)。结果表明TCS JN675228/JN675229通过多种途径调控保加利亚乳杆菌的酸适应能力,包括调控质子泵相关蛋白、经典应激休克蛋白、碳水化合物代谢、核苷酸生物合成、DNA修复转录和翻译、肽转运和降解及细胞壁生物合成。
表1 保加利亚乳杆菌CH3菌株TCS系统JN675228/JN675229靶基因
Table 1 Target genes of two-component signal transduction system JN675228/ JN675229 in L.bulgaricus CH3
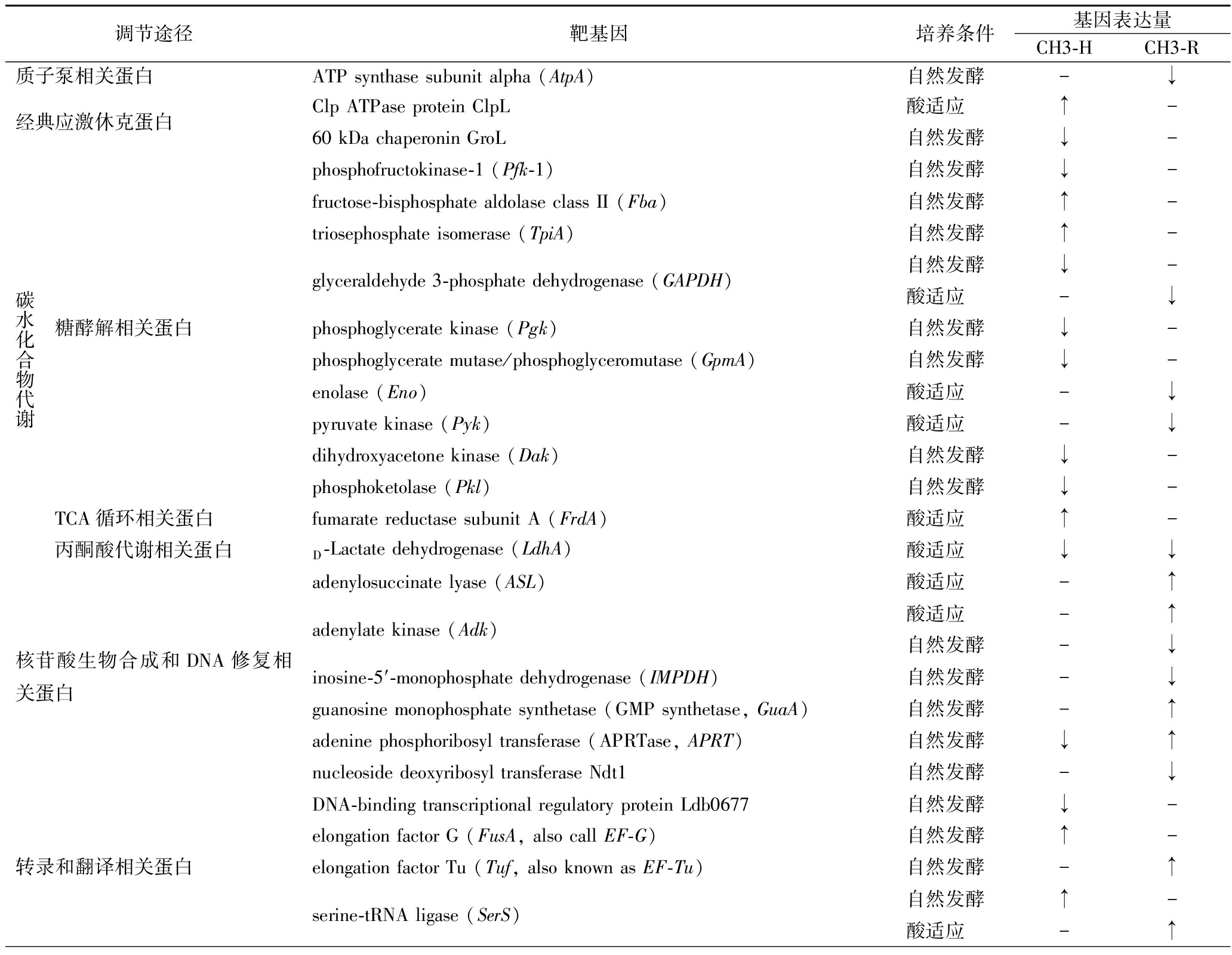
调节途径靶基因培养条件基因表达量CH3-HCH3-R质子泵相关蛋白ATP synthase subunit alpha (AtpA)自然发酵-↓经典应激休克蛋白Clp ATPase protein ClpL酸适应↑-60 kDa chaperonin GroL自然发酵↓-碳水化合物代谢糖酵解相关蛋白TCA循环相关蛋白丙酮酸代谢相关蛋白phosphofructokinase-1 (Pfk-1)fructose-bisphosphate aldolase class II (Fba)triosephosphate isomerase (TpiA)glyceraldehyde 3-phosphate dehydrogenase (GAPDH)phosphoglycerate kinase (Pgk)phosphoglycerate mutase/phosphoglyceromutase (GpmA)enolase (Eno)pyruvate kinase (Pyk)dihydroxyacetone kinase (Dak)phosphoketolase (Pkl)fumarate reductase subunit A (FrdA)D-Lactate dehydrogenase (LdhA)自然发酵↓-自然发酵↑-自然发酵↑-自然发酵↓-酸适应-↓自然发酵↓-自然发酵↓-酸适应-↓酸适应-↓自然发酵↓-自然发酵↓-酸适应↑-酸适应↓↓核苷酸生物合成和DNA修复相关蛋白adenylosuccinate lyase (ASL)adenylate kinase (Adk)inosine-5′-monophosphate dehydrogenase (IMPDH)guanosine monophosphate synthetase (GMP synthetase, GuaA)adenine phosphoribosyl transferase (APRTase, APRT)nucleoside deoxyribosyl transferase Ndt1酸适应-↑酸适应-↑自然发酵-↓自然发酵-↓自然发酵-↑自然发酵↓↑自然发酵-↓转录和翻译相关蛋白DNA-binding transcriptional regulatory protein Ldb0677elongation factor G (FusA, also call EF-G)elongation factor Tu (Tuf, also known as EF-Tu)serine-tRNA ligase (SerS)自然发酵↓-自然发酵↑-自然发酵-↑自然发酵↑-酸适应-↑
续表1
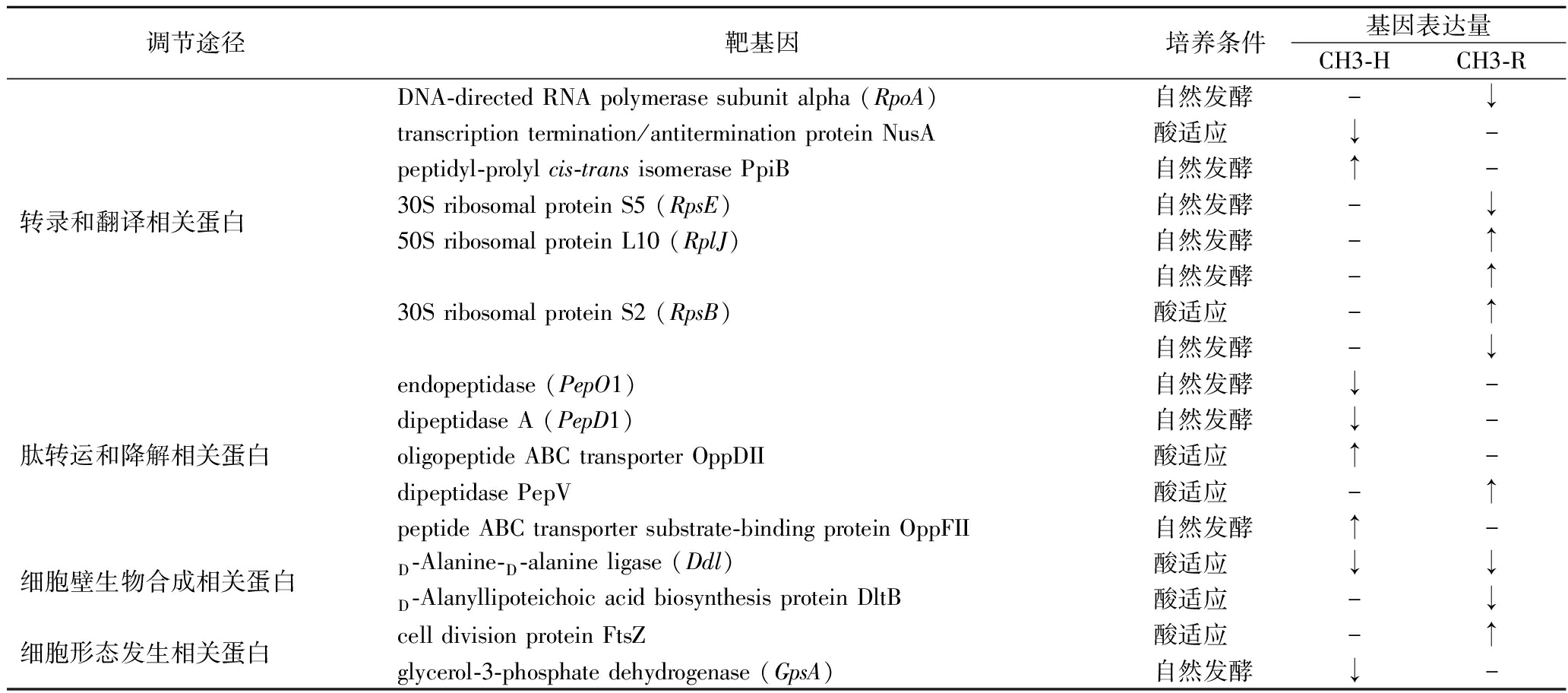
调节途径靶基因培养条件基因表达量CH3-HCH3-R转录和翻译相关蛋白DNA-directed RNA polymerase subunit alpha (RpoA)transcription termination/antitermination protein NusApeptidyl-prolyl cis-trans isomerase PpiB30S ribosomal protein S5 (RpsE)50S ribosomal protein L10 (RplJ)30S ribosomal protein S2 (RpsB)自然发酵-↓酸适应↓-自然发酵↑-自然发酵-↓自然发酵-↑自然发酵-↑酸适应-↑自然发酵-↓肽转运和降解相关蛋白endopeptidase (PepO1)自然发酵↓-dipeptidase A (PepD1)自然发酵↓-oligopeptide ABC transporter OppDII酸适应↑-dipeptidase PepV酸适应-↑peptide ABC transporter substrate-binding protein OppFII自然发酵↑-细胞壁生物合成相关蛋白D-Alanine-D-alanine ligase (Ddl)酸适应↓↓D-Alanyllipoteichoic acid biosynthesis protein DltB酸适应-↓细胞形态发生相关蛋白cell division protein FtsZ酸适应-↑glycerol-3-phosphate dehydrogenase (GpsA)自然发酵↓-
注:CH3-H表示JN675228敲除突变株,CH3-R表示JN675229敲除突变株;↑表示显著上调;-表示无显著变化;↓表示显著下调
1.2 调控细胞自溶
乳酸菌自溶对乳制品加工至关重要,在干酪成熟过程,乳酸菌通过自溶将胞内肽酶释放到干酪中,降解氨基酸产生风味物质[12];而在生产酸奶时乳酸菌自溶则会降低起始菌株的活细胞数[13]。PANG等[14]通过生物信息学方法对保加利亚乳杆菌BAA-365中TCS进行功能预测(表2),结果显示BAA-365中5对TCS功能与细胞自溶无关,2个功能未知。对其RR(WP_011543855.1和WP_011677872.1)编码基因敲除,细胞自溶评估实验表明WP_011543855.1编码基因敲除对菌株的自溶率和菌落总数没有明显影响,而WP_011677872.1编码基因敲除使菌株自溶率显著降低,菌落总数显著上升,基因回补后差异消除。酵母双杂交实验证实WP_011677872.1和WP_011677871.1存在直接相互作用。因此,TCS WP_011677872.1/WP_011677871.1调控保加利亚乳杆菌细胞自溶。此外,这对TCS还参与群体感应信号分子肽II调控细胞密度[15]。
表2 保加利亚乳杆菌BAA-365菌株TCS系统功能预测
Table 2 Functional prediction of two-component signal transduction system in L.bulgaricus BAA-365
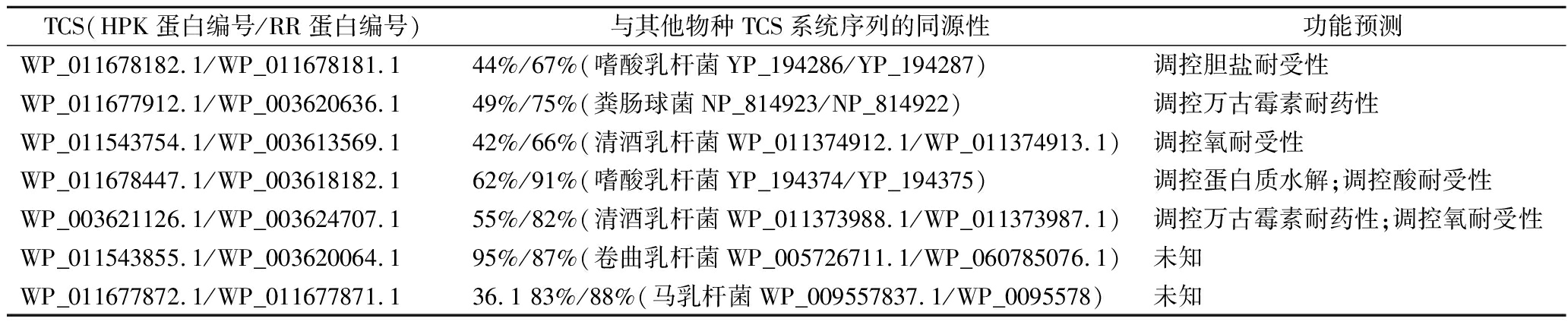
TCS(HPK蛋白编号/RR蛋白编号)与其他物种TCS系统序列的同源性功能预测WP_011678182.1/WP_011678181.144%/67%(嗜酸乳杆菌YP_194286/YP_194287)调控胆盐耐受性WP_011677912.1/WP_003620636.149%/75%(粪肠球菌NP_814923/NP_814922)调控万古霉素耐药性WP_011543754.1/WP_003613569.142%/66%(清酒乳杆菌WP_011374912.1/WP_011374913.1)调控氧耐受性WP_011678447.1/WP_003618182.162%/91%(嗜酸乳杆菌YP_194374/YP_194375)调控蛋白质水解;调控酸耐受性WP_003621126.1/WP_003624707.155%/82%(清酒乳杆菌WP_011373988.1/WP_011373987.1)调控万古霉素耐药性;调控氧耐受性WP_011543855.1/WP_003620064.195%/87%(卷曲乳杆菌WP_005726711.1/WP_060785076.1)未知WP_011677872.1/WP_011677871.136.1 83%/88%(马乳杆菌WP_009557837.1/WP_0095578)未知
2 嗜热链球菌TCS
嗜热链球菌是一种同型发酵乳酸菌,在适应牛奶环境过程中,嗜热链球菌失去了许多与毒力和糖利用相关的基因,但保留了参与牛奶发酵所需的氮代谢和乳糖利用等功能基因[16]。另外,嗜热链球菌的代谢产物胞外多糖(exopolysaccharide, EPS)不仅具有降低胆固醇、抗肿瘤和促进肠道黏附等生理功能,还能作为食品添加剂和稳定剂显著改善食品物性[17]。产EPS与核苷酸糖代谢和EPS生物合成相关基因的高转录水平相关[18],精氨酸代谢调控蛋白ArgR能特异性结合eps基因簇启动子,负调控EPS合成[19]。
不同嗜热链球菌菌株具有6~8对完整TCS,例如嗜热链球菌LMD-9菌株具有6对完整TCS(TCS02、04、05、06、07和09)及2个孤立RR(RR01和RR08),其中TCS05对嗜热链球菌生长至关重要。转录组学研究表明嗜热链球菌LMD-9菌株所有RR基因在牛奶发酵过程中表达,但其表达水平不同[20]。
2.1 调控生产特性
牛奶发酵中嗜热链球菌产生乳酸,使环境pH迅速下降,有助于牛奶酸化;嗜热链球菌合成EPS,有助于提高乳制品的黏度和改善质地[21];嗜热链球菌蛋白水解活性与生长能力、牛奶酸化速率和芳香化合物形成密切相关。HU等[22]从内蒙古地区传统酸奶样品中分离鉴定出22株嗜热链球菌,根据已公布的嗜热链球菌TCS序列信息,设计引物扩增22株嗜热链球菌的TCS。结果表明,22株嗜热链球菌均具有8对TCS,但不同菌株的TCS基因序列存在差异。聚类分析发现菌株的不同生产特性与TCS差异存在相关性,其中系统发育树属于Ⅰ-1组和主成分分析属于A簇菌株,它们具有较高的牛奶酸化活性、EPS合成能力、蛋白水解活性等相似的生产特性;系统发育树属于Ⅲ组和主成分分析属于A簇菌株,TCS数量和基因序列完全相同,表现出相似的碳水化合物利用能力及牛奶酸化活性;而系统发育树属于Ⅳ-2组和主成分分析属于B簇的菌株,与其他菌株相比具有独特的TCS序列,表现出显著不同的生产特性。这些结果为进一步研究嗜热链球菌TCS与生产特性的关系奠定基础。
2.2 调控细菌素生产
细菌素是细菌在代谢过程中通过核糖体途径合成的一类具有抗菌活性的多肽类物质。乳酸菌细菌素具有良好的生物安全性、稳定性及适应工业化生产等优点[23],在食品防腐保鲜方面应用广泛。HOLS等[20]在嗜热链球菌基因组中发现II类细菌素基因簇,与肺炎链球菌类细菌素肽(bacteriocin-like peptide, blp)基因簇blpSp相似,命名为嗜热链球菌blpSt。LMD-9 blpSt基因簇包含23个基因,分为6个操纵子,包括肽转运基因和信息素基因blpABCSt,TCS基因blpRHSt(即TCS09,BlpHSt和BlpRSt分别为HPK和RR),细菌素前体和免疫基因blpDSt-orf1、blpUSt-orf3和blpE-FSt及功能未知基因blpG-XSt。为研究blpSt基因簇功能,FONTAINE等[24]构建blpSt敲除突变株,在以BlpCSt为前体的成熟信息素诱导下通过spot-on-lawn法和覆盖法观察其细菌素生产和免疫力表型(表3),结果表明blpSt基因簇与细菌素生产和免疫调节有关。通过Northern印迹杂交进行转录组分析发现,blpSt启动子中存在直接重复序列ACCATTCGGG和ACTTTTTGGG,作为BlpRSt假定结合位点;细菌素生产受BlpCSt浓度和生长阶段调控,表现出群体感应特征。这些结果证明blpSt基因簇调控细菌素生产,通过信息素介导的群体感应激活TCS09完成调控。FONTAINE等[25]后续证明blpSt基因簇编码嗜热链球菌素9多肽细菌素系统,能抑制链球菌、粪肠球菌、乳酸杆菌和部分李斯特菌等大多数与嗜热链球菌密切相关的细菌生长。另外,在嗜热链球菌与保加利亚乳杆菌共培养条件下,blpRSt表达量增加6倍,表明TCS09的表达显著受保加利亚乳杆菌的影响[20]。
表3 嗜热链球菌LMD-9及其敲除突变株表型
Table 3 Phenotype of S.thermophilus LMD-9 derivatives

菌株细菌素生产免疫力LMD-9野生型菌株++blpRSt敲除突变株--blpHSt敲除突变株--blpRSt-blpHSt敲除突变株--blpDSt-blpXSt敲除突变株--blpDSt-blpFSt敲除突变株--blpGSt-blpXSt敲除突变株++blpBSt敲除突变株++blpASt-blpBSt敲除突变株-+
注:+表示阳性;-表示阴性
2.3 调控杆菌肽耐药性
杆菌肽是由芽孢杆菌产生的一种新型环状肽抗生素[26],对革兰氏阳性细菌和阴性球菌及螺旋体均有杀菌作用,主要作用机制是抑制细菌肽聚糖合成,通过与膜受体十一烯丙基焦磷酸结合,阻止细胞壁的生物合成,抑制其去磷酸化,从而破坏十一烯丙基磷酸的再生[27]。因其具有广谱抗菌活性、禽畜低吸收率、快速排泄率、高安全性和不易产生耐药性等特点,在饲料行业被广泛用作饲料添加剂[28]。嗜热链球菌LMD-9中TCS06和TCS07在基因组上位置接近,TCS06同源物常在乳酸杆菌中被发现[29],TCS07及其上游潜在的ABC转运蛋白与枯草芽孢杆菌解毒模块BceRS/AB系统同源,推测TCS06和TCS07与嗜热链球菌杆菌肽耐药性相关。为验证此推测,THEVENARD[30]等采用qPCR方法评估TCS06和TCS07附近基因在杆菌肽耐药中表达变化,发现rr07及其上游ABC转运蛋白编码基因STER_1 308-1307表达量上调,并且RR07显著诱导STER_1308表达。生长实验发现,rr07和STER_1 308-1307敲除突变株对杆菌肽更敏感。这些结果表明TCS07及其ABC转运蛋白构成一个解毒模块,STER_1 308-1307在杆菌肽的泵出中发挥重要作用,且受TCS07正向调控。鼠李糖-葡萄糖多糖(rhamnose-glucose polysaccharide,RGP)合成途径中,dTDP-4-脱氢鼠李糖3,5-差向异构酶RmlC催化D-葡萄糖-1-磷酸生成dTDP -L-鼠李糖,葡萄糖基转移酶RgpI控制RGP鼠李糖骨架上形成葡萄糖分支。在杆菌肽胁迫下,野生型菌株rmlC和rgpI表达量显著上调,rr06敲除菌株中3种RGP聚合酶编码基因rgpA、rgpB和rgpC表达量也显著上调。rmlC敲除突变株对杆菌肽的敏感性显著提高,电镜观察发现其细胞膜形态异常。以上结果表明TCS06可能通过调控RGP合成和细胞膜形态影响嗜热链球菌的杆菌肽耐药性。
2.4 调控抗逆性
嗜热链球菌Sfi39发酵性能优良,但抗逆性较差[31]。Sfi39对数生长期细胞对酸和氧敏感,但对渗透压和热较为耐受;酸或热适应均使Sfi39耐热性显著提升,耐酸性略微提升,而氧耐受性不受酸或热适应的影响[32]。ZOTTA等[33]构建covR同源基因rr01敲除突变株比较其耐受表型。与野生型菌株相比,rr01敲除菌株对数生长期和稳定期细胞的酸和氧耐受性均显著提高,热和渗透耐受性均显著降低;与未经适应的rr01敲除株对数生长期细胞相比,酸或热适应均使其酸耐受性显著提高,渗透耐受性显著降低,酸适应使热耐受性略微提高,氧耐受性无明显变化,热适应使热耐受性和氧耐受性显著降低;rr01敲除突变株的SDS-PAGE图谱与野生型相似,但主成分分析表明两者存在显著差异。以上结果表明rr01敲除使热和渗透耐受性降低,但机制仍有待研究。
3 总结与展望
作为细菌中最常见的信号机制,TCS一直是研究热点。许多细菌TCS已有深入研究,尤其是致病菌TCS凭借其基因保守性特点,成为新兴安全高效的药物作用靶点。但乳酸菌TCS的研究偏少,例如保加利亚乳杆菌和嗜热链球菌中多数TCS功能仍不清晰,而植物乳杆菌和嗜酸乳杆菌等其他常见乳酸菌的TCS功能知之甚少,且乳酸菌TCS研究主要集中在RR,对HPK鲜有涉及。制约乳酸菌TCS研究的关键因素之一是其基因编辑方法缺乏。根据已有的研究表明,TCS与乳酸菌的生长和功能活性息息相关。因此,研究乳酸菌TCS的调控机制,有助于利用传统选育、分子生物学和代谢工程等方法改善乳酸菌的生长和性能,开发出更加优良的酸奶发酵剂菌种。
[1] MASOOD M I, QADIR M I, SHIRAZI J H, et al.Beneficial effects of lactic acid bacteria on human beings[J].Critical Reviews in Microbiology, 2011, 37(1):91-98.
[2] MOKOENA M P, MUTANDA T, OLANIRAN A O.Perspectives on the probiotic potential of lactic acid bacteria from African traditional fermented foods and beverages[J].Food & Nutrition Research, 2016, 60:29630.
[3] 苏敦, 任发政, 刘松玲, 等.菌株发酵特性研究及复合酸奶发酵剂的筛选[J].中国奶牛, 2017(9):50-57.
SU D, REN F Z, LIU S L, et al.Study of bacterial strains fermentation characteristics and development of yoghurt starter[J].China Dairy Cattle, 2017(9):50-57.
[4] 杜心恬, 宋馨, 刘欣欣, 等.细菌胞外多糖生物合成转录调控因子研究进展[J].微生物学通报, 2021, 48(2):573-581.
DU X T, SONG X, LIU X X, et al.Advances in transcription regulators of bacterial exopolysaccharides biosynthesis[J].Microbiology China, 2021, 48(2):573-581.
[5] ZHANG D F, YE J Z, DAI H H, et al.Identification of ethanol tolerant outer membrane proteome reveals OmpC-dependent mechanism in a manner of EnvZ/OmpR regulation in Escherichia coli [J].Journal of Proteomics, 2018, 179:92-99.
[6] ZHENG C K, LI L Z, GE H J, et al.Role of two-component regulatory systems in the virulence of Streptococcus suis[J].Microbiological Research, 2018, 214:123-128.
[7] YUAN J, JIN F, GLATTER T, et al.Osmosensing by the bacterial PhoQ/PhoP two-component system[J].Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(50):E10792-E10798.
[8] VAN DE GUCHTE M, PENAUD S, GRIMALDI C, et al.The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution[J].Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(24):9 274-9 279.
[9] MAKAROVA K, SLESAREV A, WOLF Y, et al.Comparative genomics of the lactic acid bacteria[J].Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(42):15 611-15 616.
[10] WANG C, CUI Y H, QU X J.Identification of proteins regulated by acid adaptation related two component system HPK1/RR1 in Lactobacillus delbrueckii subsp.bulgaricus[J].Archives of Microbiology, 2018, 200(9):1 381-1 393.
[11] CUI Y H, LIU W, QU X J, et al.A two component system is involved in acid adaptation of Lactobacillus delbrueckii subsp.bulgaricus[J].Microbiological Research, 2012, 167(5):253-261.
[12] 宋雪梅, 张炎, 张卫兵, 等.成熟温度对牦牛乳硬质干酪中乳酸菌自溶及氨肽酶活性的影响[J].食品与发酵工业, 2016, 42(5):38-43.
SONG X M, ZHANG Y, ZHANG W B, et al.Effect of ripening temperature on the autolysis and aminopeptidase activity of Lactococcus lactis in hard cheese from yak milk[J].Food and Fermentation Industries, 2016, 42(5):38-43.
[13] PANG X Y, CUI W M, LIU L, et al.Gene knockout and overexpression analysis revealed the role of N-acetylmuramidase in autolysis of Lactobacillus delbrueckii subsp.bulgaricus ljj-6[J].PLoS One, 2014, 9(8):e104829.
[14] PANG X Y, ZHANG S W, LU J, et al.Identification and functional validation of autolysis-associated genes in Lactobacillus bulgaricus ATCC BAA-365[J].Frontiers in Microbiology, 2017, 8:1367.
[15] PANG X Y, LIU C P, LYU P C, et al.Identification of quorum sensing signal molecule of Lactobacillus delbrueckii subsp.bulgaricus[J].Journal of Agricultural and Food Chemistry, 2016, 64(49):9 421-9 427.
[16] BOLOTIN A, QUINQUIS B, RENAULT P, et al.Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus[J].Nature Biotechnology, 2004, 22(12):1 554-1 558.
[17] XIONG Z Q, KONG L H, MENG H L, et al.Comparison of gal-lac operons in wild-type galactose-positive and-negative Streptococcus thermophilus by genomics and transcription analysis[J].Journal of Industrial Microbiology and Biotechnology, 2019, 46(5):751-758.
[18] XIONG Z Q, KONG L H, LAI P F H, et al.Genomic and phenotypic analyses of exopolysaccharide biosynthesis in Streptococcus thermophilus S-3[J].Journal of Dairy Science, 2019, 102(6):4 925-4 934.
[19] 潘晖, 刘欣欣, 孔令慧, 等.精氨酸代谢调控蛋白ArgR调控嗜热链球菌胞外多糖合成[J].微生物学报, 2020, 60(11):2 412-2 422.
PAN H, LIU X X, KONG L H, et al.Arginine regulator ArgR regulates exopolysaccharides biosynthesis of Streptococcus thermophilus[J].Acta Microbiologica Sinica, 2020, 60(11):2 412-2 422.
[20] HOLS P, HANCY F, FONTAINE L, et al.New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics[J].FEMS Microbiology Reviews, 2005, 29(3):435-463.
[21] MENDE S, ROHM H, JAROS D.Influence of exopolysaccharides on the structure, texture, stability and sensory properties of yoghurt and related products[J].International Dairy Journal, 2016, 52:57-71.
[22] HU T, ZHANG Y S, CUI Y H, et al.Technological properties assessment and two component systems distribution of Streptococcus thermophilus strains isolated from fermented milk[J].Archives of Microbiology, 2018, 200(4):567-580.
[23] CLEVELAND J, MONTVILLE T J, NES I F, et al.Bacteriocins:safe, natural antimicrobials for food preservation[J].International Journal of Food Microbiology, 2001, 71(1):1-20.
[24] FONTAINE L, BOUTRY C, GUÉDON E, et al.Quorum-sensing regulation of the production of blp bacteriocins in Streptococcus thermophilus[J].Journal of Bacteriology, 2007, 189(20):7 195-7 205.
[25] FONTAINE L, HOLS P.The inhibitory spectrum of thermophilin 9 from Streptococcus thermophilus LMD-9 depends on the production of multiple peptides and the activity of BlpG(St), a thiol-disulfide oxidase[J].Applied and Environmental Microbiology, 2008, 74(4):1 102-1 110.
[26] WANG D, WANG Q, QIU Y M, et al.Untangling the transcription regulatory network of the bacitracin synthase operon in Bacillus licheniformis DW2[J].Research in Microbiology, 2017, 168(6):515-523.
[27] STONE K J, STROMINGER J L.Mechanism of action of bacitracin:Complexation with metal ion and C55-isoprenyl pyrophosphate[J].PNAS, 1971, 68(12):3 223-3 227.
[28] LI Y, WU F, CAI D B, et al.Enhanced production of bacitracin by knocking out of amino acid permease gene yhdG in Bacillus licheniformis DW2[J]. Chinese Journal of Biotechnology, 2018, 34(6):916-927.
[29] THEVENARD B, RASOAVA N, FOURCASSIÉ P, et al.Characterization of Streptococcus thermophilus two-component systems:In silico analysis, functional analysis and expression of response regulator genes in pure or mixed culture with its yogurt partner, Lactobacillus delbrueckii subsp.bulgaricus[J].International Journal of Food Microbiology, 2011, 151(2):171-181.
[30] THEVENARD B, BESSET C, CHOINARD S, et al.Response of S.thermophilus LMD-9 to bacitracin:Involvement of a BceRS/AB-like module and of the rhamnose-glucose polysaccharide synthesis pathway[J].International Journal of Food Microbiology, 2014, 177:89-97.
[31] LEMOINE J, CHIRAT F, WIERUSZESKI J M, et al.Structural characterization of the exocellular polysaccharides produced by Streptococcus thermophilus SFi39 and SFi12[J].Applied and Environmental Microbiology, 1997, 63(9):3 512-3 518.
[32] ZOTTA T, RICCIARDI A, CIOCIA F, et al.Diversity of stress responses in dairy thermophilic streptococci[J].International Journal of Food Microbiology, 2008, 124(1):34-42.
[33] ZOTTA T, ASTERINOU K, ROSSANO R, et al.Effect of inactivation of stress response regulators on the growth and survival of Streptococcus thermophilus Sfi39[J].International Journal of Food Microbiology, 2009, 129(3):211-220.