S-腺苷甲硫氨酸(S-adenosylmethionine,SAM)是生物体中主要的甲基供体。除了甲基供体外,还参与转氨丙基和转硫基反应。SAM具有重要的药学意义,已被用于治疗多种疾病,包括阿尔茨海默病、肝脏疾病、骨关节炎和抑郁症[1-4],市场需求量巨大。SAM合成方法有化学合成法、发酵法和酶促转化法。化学合成法合成的SAM收率低,且大多数包含有非活性异构体,反应条件苛刻后期难以分离纯化,因此难以获得大规模的应用[5-6]。发酵法是目前SAM工业化生产的主要途径,该方法简单、易于控制、扩大培养和安全无污染,但是最大的缺点是L-甲硫氨酸转化率低、SAM产量较低[7]。酶促转化法是底物L-甲硫氨酸和ATP在SAM合酶催化下直接生成SAM,反应过程需要K+和Mg2+参与,该方法最大的优点是所生成的产物大多数是具有生物活性的(S,S)-SAM,因此成为近年来工业生产SAM的研究热点。
SAM合酶是酶促转化法合成SAM的关键酶,已从多种生物中分离出来并进行表征,包括大肠杆菌[8]、枯草芽孢杆菌[9]、大鼠[10]、人[11]、詹氏甲烷球菌[12]、极端嗜热古生菌[13]、酿酒酵母[14]等。酿酒酵母是公认的SAM产量最高的微生物,由两种同工酶(SAM合酶1和SAM合酶2)催化合成,这两种同工酶氨基酸同源性达到92%,SAM合酶1存在底物抑制,SAM合酶2不存在底物抑制现象,且酶活力要高于SAM合酶1[15-16]。大肠杆菌来源的SAM合酶存在高产物抑制现象,SAM可与SAM合酶形成惰性的酶复合物,从而影响底物ATP与L-甲硫氨酸之间的相互作用[17]。
目前,工业上常用酿酒酵母(Saccharomyces cerevisiae)和毕赤酵母(Pichia pastoris)进行高密度发酵生产SAM[18]。当发酵培养基中L-甲硫氨酸含量很高时,酵母对L-甲硫氨酸的吸收会受到限制[19],SAM产量通常低于15 g/L。为解决生产中的限制因素需要制定策略,以提高SAM的可持续工业化生产。在这项研究中,我们在大肠杆菌中异源表达了蜡状芽孢杆菌和谷氨酸棒状杆菌来源的SAM合酶,针对SAM合成菌株全细胞催化合成体系条件进行了优化。有研究发现对甲苯磺酸钠的添加可改善SAM合酶的产物抑制[20-22],因此,为了改善SAM合酶在产物积累过程中的反馈抑制,在全细胞转化体系中添加了对甲苯磺酸钠,改善了SAM对SAM合酶的反馈抑制,进一步提高了SAM含量。
1 材料与方法
1.1 材料
1.1.1 菌株与质粒
本研究中所用到的质粒pETDuet1、菌株蜡状芽孢杆菌(Bacillus cereus)、谷氨酸棒杆菌(Corynebacterium glutamicum)、大肠杆菌(Escherichia coli) BL21(DE3)均为本实验室保藏。
1.1.2 实验试剂
BamH Ⅰ、EcoR Ⅰ、Hind Ⅲ和Not Ⅰ限制性快切酶、10 000 bp核酸Marker和蛋白Marker,大连宝生物公司;高保真酶、同源重组酶克隆试剂盒,南京诺维赞生物科技有限公司;质粒提取试剂盒、DNA胶回收试剂盒,上海捷瑞生物工程有限公司;SAM标准品、ATP,Sigma公司,L-甲硫氨酸及培养基原料等,国药集团。
1.1.3 培养基
LB培养基:10 g/L蛋白胨、5 g/L酵母提取物和10 g/L氯化钠,固体培养基按1.5%~2.0%(质量分数)添加琼脂粉,根据需要加入相应浓度抗生素。
1.2 实验方法
1.2.1 重组菌株E.coli BL21/pETDuet1-BcmetK和E.coli BL21/pETDuet1-CgmetK的构建
本研究所用引物如表1所示。分别以蜡状芽孢杆菌和谷氨酸棒杆菌基因组为模板,以pETDuet1-BcmetK-F/R和pETDuet1-CgmetK-F/R为引物进行PCR扩增,PCR产物纯化后得到的BcmetK和CgmetK基因片段分别与经BamH Ⅰ/EcoR Ⅰ和Hind Ⅲ/NotⅠ双酶切线性化的pETDuet1质粒进行同源重组连接,连接产物分别化转至E.coli BL21感受态细胞,37 ℃摇床培养2 h后,涂布至含50 μg/mL氨苄青霉素的LB平板上,37 ℃培养箱倒置培养8~12 h。挑取转化子进行菌落PCR验证,验证正确的转化子进行培养并提取重组质粒送苏州金唯智生物科技有限公司进行测序分析,测序正确的重组菌株命名为E.coli BL21/pETDuet1-BcmetK和E.coli BL21/pETDuet1-CgmetK。
表1 本研究中使用的引物
Table 1 Primers used in this research
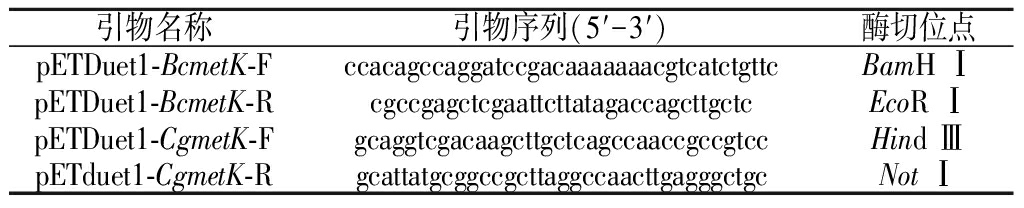
引物名称引物序列(5′-3′)酶切位点pETDuet1-BcmetK-FccacagccaggatccgacaaaaaaacgtcatctgttcBamH ⅠpETDuet1-BcmetK-RcgccgagctcgaattcttatagaccagcttgctcEcoR ⅠpETDuet1-CgmetK-FgcaggtcgacaagcttgctcagccaaccgccgtccHind ⅢpETduet1-CgmetK-RgcattatgcggccgcttaggccaacttgagggctgcNot Ⅰ
1.2.2 SAM合酶的表达及纯化
将冻管保存的E.coli BL21/pETDuet1-BcmetK和E.coli BL21/pETDuet1-CgmetK重组菌株分别在含50 μg/mL氨苄青霉素的LB平板上划线活化,挑取单菌落转接10 mL含50 μg/mL氨苄青霉素的LB液体培养基,37 ℃、200 r/min下培养8~12 h后,按10%接种量转接50 mL含相同浓度的LB液体培养基,37 ℃、200 r/min下培养2 h后,添加异丙基-β-D硫代半乳糖苷(isopropyl-β-D-thiogalactopyranoside,IPTG)(终浓度为0.5 mmol/L),然后于25 ℃、200 r/min条件下培养18 h,以诱导SAM合酶的表达。对照菌株E.coli BL21/pETDuet1按照相同方法进行诱导表达。
之后,在4 ℃、10 000 r/min下离心10 min收集细胞,弃去培养基,细胞菌体用5 mL Tris-HCl缓冲液(50 mmol/L,pH 7.0)洗2次。最后,将细胞菌体悬浮于5 mL Tris-HCl缓冲液(50 mmol/L,pH 7.0)中并用超声破碎仪进行破碎,破碎液在4 ℃下离心20 min,收集上清液,用于后续的SDS-PAGE分析和SAM合酶的纯化。粗酶液上清用0.2 μm滤膜过滤处理后,经Ni-NTA亲和柱纯化后收集SAM合酶纯酶,测定酶活力水平。
1.2.3 SAM合酶的酶活力测定
酶活力测定体系[23]:10 mmol/L ATP、10 mmol/L L-甲硫氨酸、50 mmol/L K2SO4、20 mmol/L MgSO4、100 mmol/L Tris-HCl, pH 7.0,37 ℃下预热5 min后分别加入5 μL BcMAT(蜡状芽孢杆菌来源的SAM合酶)和CgMAT(谷氨酸棒杆菌来源的SAM合酶)纯酶来启动反应,30 min后,测量从ATP释放的无机磷酸盐浓度。反应体系中加入100 μL的孔雀绿钼酸盐溶液,孵育5 min后测量620 nm处的吸光值。以不添加SAM合酶的纯酶体系作为阴性对照,标准磷酸盐溶液作为阳性对照。
1.2.4 全细胞生物催化剂的制备
首先,将冻管保存的E.coli BL21/pETDuet1-BcmetK和E.coli BL21/pETDuet1-CgmetK重组菌株分别在含50 μg/mL氨苄青霉素的LB平板上划线活化,挑取单菌落转接10 mL含50 μg/mL氨苄青霉素的LB培养基,37 ℃、200 r/min下培养8~12 h后,按10%接种量转接100 mL含相同浓度氨苄青霉素的LB培养基,37 ℃、180 r/min下培养2 h后,添加IPTG(终浓度0.5 mmol/L),25 ℃、200 r/min下培养18 h,以诱导SAM合酶表达。之后,在4 ℃、10 000 r/min下离心10 min收集细胞,弃去培养基,用Tris-HCl缓冲液(50 mmol/L,pH 7.0)洗涤菌体2次,即获得全细胞生物催化剂。
1.2.5 全细胞催化条件优化
为了优化全细胞生物转化合成SAM的条件,将离心收集的过表达SAM合酶的细胞菌体用Tris-HCl缓冲液(50 mmol/L,pH 7.0)洗涤菌体2次后并悬浮。为了研究全细胞生物催化的最佳温度,将反应体系分别在25、30、35、40、45、50 ℃、180 r/min的旋转振荡器中运行。对于pH的优化,反应体系在初始pH为5.0~9.0(pH间隔1.0)下反应。为了确定生物催化剂的最佳浓度,反应体系分别使用0.2%~1.6%(体积分数,间隔0.2%)的OD600值约9.0的重组菌株全细胞菌体悬浮液作为生物催化剂进行催化合成SAM。最后,测试了不同浓度10~60 mmol/L的底物L-甲硫氨酸和ATP对全细胞催化合成SAM的影响,以确定最优底物浓度。
1.2.6 反馈抑制实验
实验组反应体系:50 mmol/L L-甲硫氨酸、适当浓度ATP(BcMAT:50 mmol/L、CgMAT:40 mmol/L)、50 mmol/L MgSO4、100 mmol/L K2SO4、200~1 000 mmol/L对甲苯磺酸钠(sodium p-toluenesulfonate,pTSoNa)、适当菌体浓度(BcMAT:1.2%(体积分数)的OD600值为9.0的细胞悬浮液、CgMAT:0.8%(体积分数)的OD600值为9.0的细胞悬浮液)。对照组则不添加对甲苯磺酸钠,其他均一致。45 ℃、各自最适pH值(BcMAT:8.0,CgMAT:8.5)下反应5 h后加入6%(体积分数)的高氯酸溶液终止反应。最后,12 000 r/min离心10 min,上清液进行HPLC检测SAM含量。
1.2.7 序列比对分析
将从Uniprot 数据库查询到的BcMAT(登录号:Q816Q8)和CgMAT(登录号:Q9K5E4)蛋白序列使用NCBI的COBALT工具(https://www.ncbi.nlm.nih.gov/tools/cobalt/)进行比对分析。
1.2.8 S-腺苷甲硫氨酸的HPLC检测分析
采用C18 column (Hypersil BDS column, 4.6 mm×250 mm, 5 μm)色谱柱,流动相为0.01 mol/L甲酸铵溶液(pH 3.0),流速为0.8 mL/min,柱温为35 ℃,进样量10 μL,在紫外检测波长254 nm下检测S-腺苷甲硫氨酸含量。
2 结果与分析
2.1 不同来源SAM合酶编码基因metK的克隆及重组菌株的构建
以蜡状芽孢杆菌和谷氨酸棒杆菌基因组为模板,以pETDuet1-BcmetK-F/R和pETDuet1-CgmetK-F/R为引物进行PCR扩增获得BcmetK和CgmetK基因片段,基因大小分别为1 200 bp和1 224 bp。
胶回收纯化的BcmetK和CgmetK基因片段分别与经BamH Ⅰ/EcoRⅠ和Hind Ⅲ/Not Ⅰ双酶切线性化的pETDuet1质粒进行同源重组连接,连接产物转化E.coli BL21感受态细胞。挑取转化子进行菌落PCR验证,验证正确的转化子进行培养并提取重组质粒送苏州金唯智生物科技有限公司进行测序分析,测序正确的重组菌株命名为E.coli BL21/pETDuet1-BcmetK和E.coli BL21/pETDuet1-CgmetK。
2.2 不同来源SAM合酶的表达、纯化及酶活力测定
按1.2.2所述方法对蜡状芽胞杆菌和谷氨酸棒杆菌来源的SAM合酶进行诱导表达,SDS-PAGE结果显示,E.coli BL21/pETDuet1-BcmetK和E.coli BL21/pETDuet1-CgmetK上清液分别在43.0、44.1 kDa附近有条带加粗,即BcMAT和CgMAT成功实现了在E.coli BL21中的异源表达(图1-A)。
重组菌株E.coli BL21/pETDuet1-BcmetK和E.coli BL21/pETDuet1-CgmetK上清液的蛋白纯化结果如图1-B所示,酶活力测定结果显示,CgMAT的酶活力为2.05 U/mL,是BcMAT酶活力的1.3倍(1.53 U/mL)。
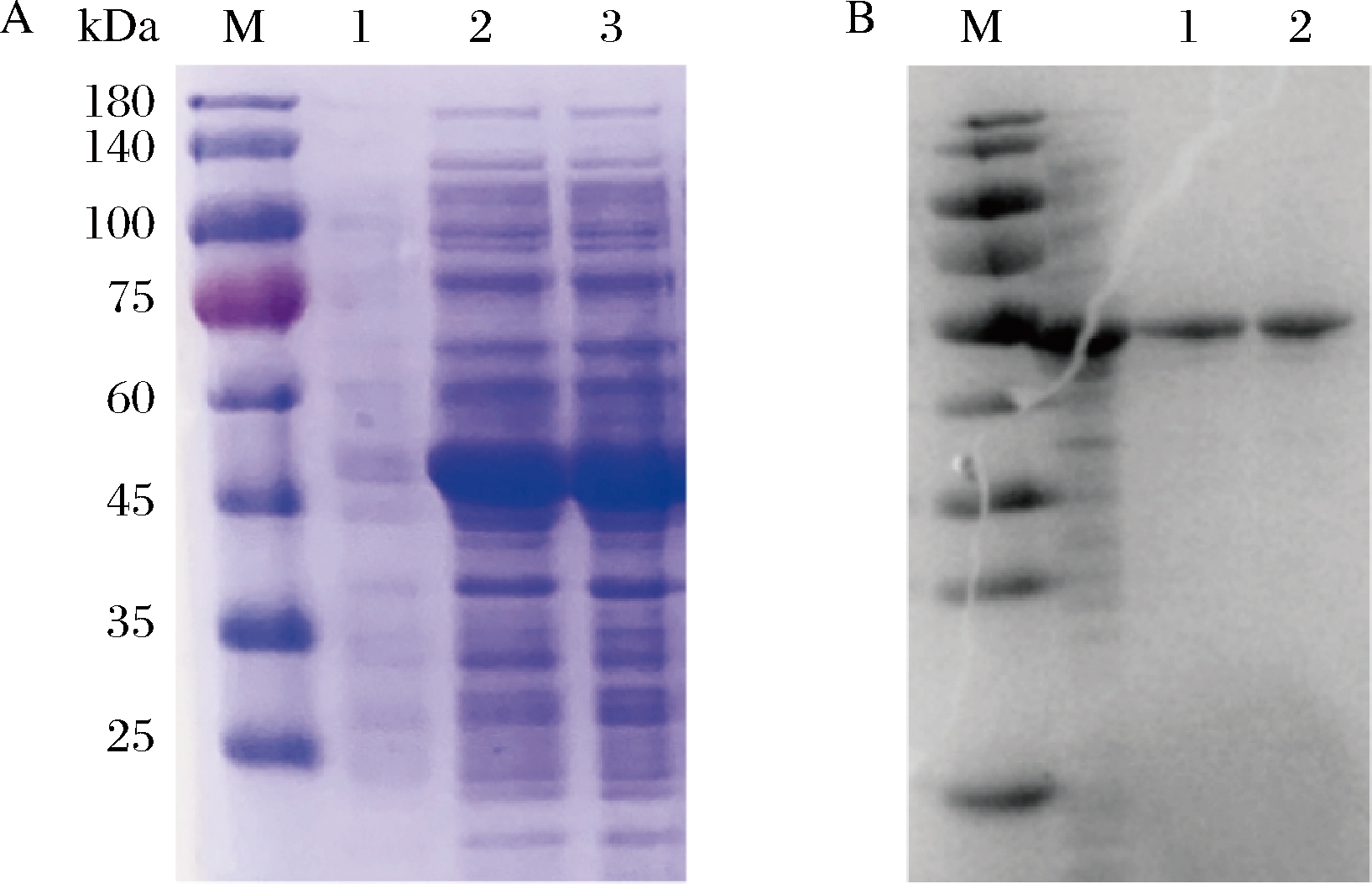
M-蛋白Marker A-异源表达结果(1-E.coli BL21/pETDuet1对照菌株细胞 破碎上清液;2-E.coli BL21/pETDuet1-BcmetK重组菌株细胞 破碎上清液;3-E.coli BL21/pETDuet1-CgmetK重组菌株细胞 破碎上清液);B-蛋白纯化结果(1-BcMAT纯酶;2-CgMAT纯酶)
图1 不同来源SAM合酶的表达及纯化
Fig.1 Expression and purification of SAM synthase from different sources
2.3 全细胞催化条件的优化
全细胞生物催化法应用于合成目标产物在合成生物学中已普及,其存在副反应最小化、产物更容易分离和更少环境问题的优点[24]。全细胞生物转化生成目标产物过程中关键酶活性和反应效率通常受反应条件的影响。温度、pH、底物浓度和菌体浓度等因素会影响目标产物合成。因此,为获得SAM的最佳产量,该研究针对全细胞催化生物转化合成SAM的温度、pH、底物浓度及菌体浓度进行了优化研究。
2.3.1 反应温度的优化
在工业中,温度非常重要,它会影响酶的稳定性、活性和最终的生产力,因此,为了确定最佳反应温度,20 mL反应体系由50 mmol/L L-甲硫氨酸、40 mmol/L ATP、50 mmol/L pH 7.0 Tris-HCl、50 mmol/L MgSO4、100 mmol/L K2SO4组成。将100 μL生物催化剂(OD600值约9.0)分别加入到反应体系中开始反应,反应体系在不同温度(25、30、35、40、45、50 ℃)下反应5 h。如图2所示,在25~45 ℃,SAM的含量随着温度升高而升高,当温度为45 ℃时,SAM的含量最高,重组菌株E.coli BL21/pETDuet1-CgmetK可转化合成205 mg/L的SAM,E.coli BL21/pETDuet1-BcmetK可转化合成189 mg/L的SAM。而温度高于45 ℃时,SAM的含量则逐渐下降。也就是说,E.coli BL21/pETDuet1-CgmetK和E.coli BL21/pETDuet1-BcmetK重组菌株的最适转化温度均为45 ℃。
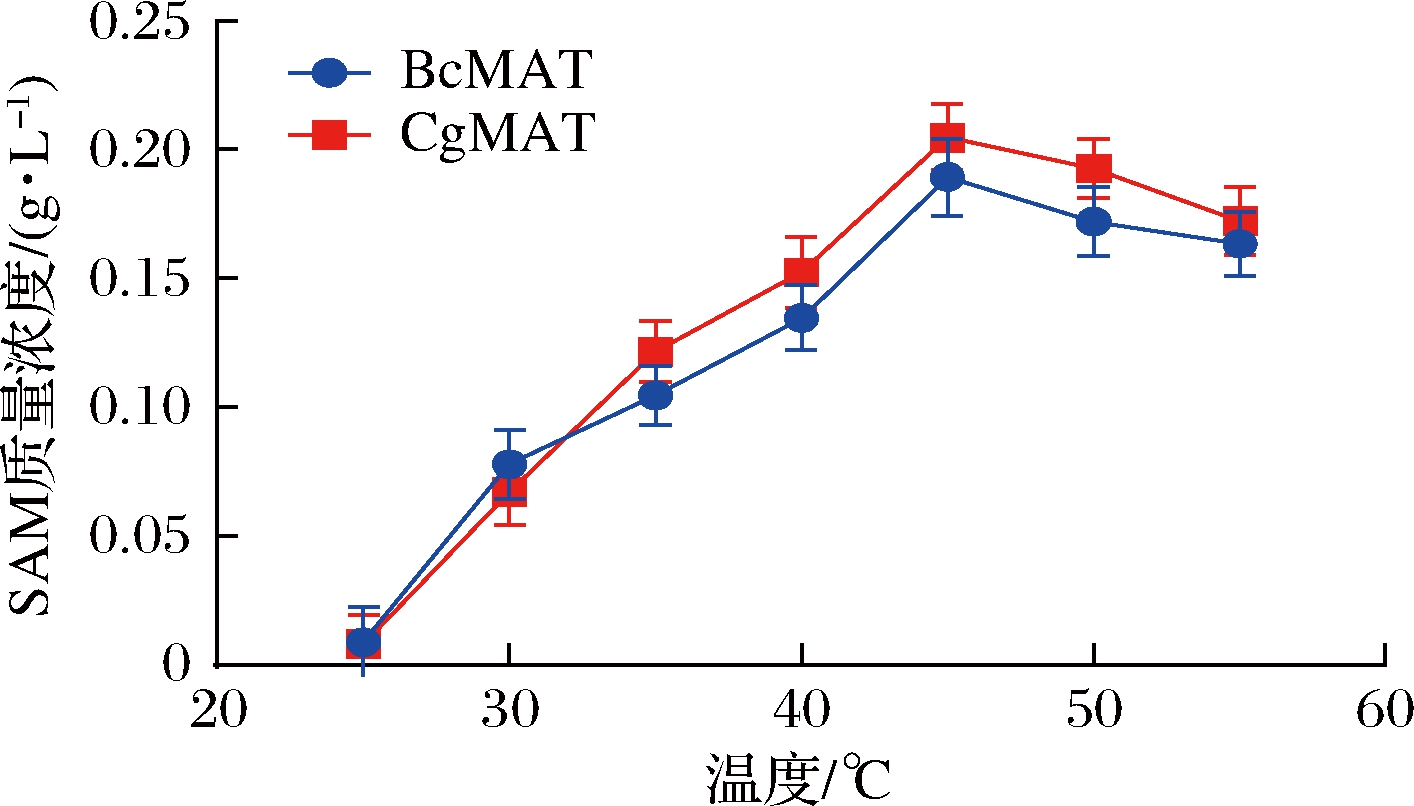
图2 反应温度对全细胞催化反应的影响
Fig.2 The effect of reaction temperature on the catalytic reaction of whole cells
2.3.2 反应pH的优化
为了确定SAM合成的最佳pH值,20 mL反应体系由50 mmol/L L-甲硫氨酸、40 mmol/L ATP、50 mmol/L MgSO4、100 mmol/L K2SO4组成,100 μL生物催化剂(OD600值约9.0)分别加入到反应体系中开始反应。全细胞转化体系的pH值控制在5.0~9.0,45 ℃下反应5 h。结果如图3所示,BcMAT和CgMAT均在pH 6.0~9.0检测到显著的酶活性。E.coli BL21/pETDuet1-BcmetK重组菌株的最佳转化pH值为8.0,此时SAM含量为232 mg/L;E.coli BL21/pETDuet1-CgmetK的最佳转化pH值为8.5,此时SAM含量为254 mg/L。
2.3.3 底物浓度的优化
底物浓度通常被视为提高全细胞生物催化效率需进行考虑调整的重要参数。底物的高初始浓度通常会抑制催化进行。为了确定重组SAM合酶进行全细胞转化的底物L-甲硫氨酸和ATP的最佳浓度,将底物L-甲硫氨酸和ATP分别设为10~60 mmol/L,转化温度和pH均使用前面优化得到的最佳值(图4)。
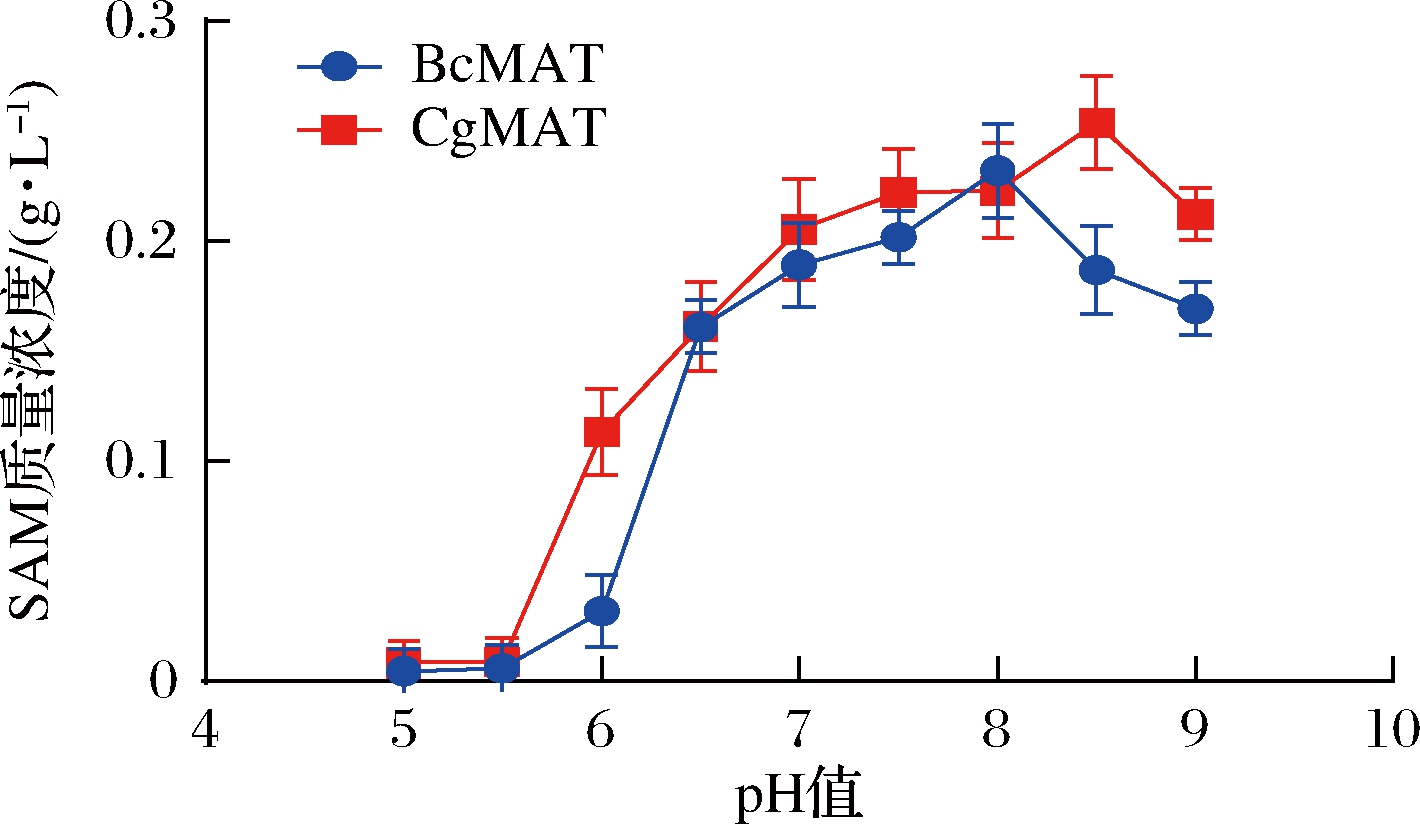
图3 反应pH对全细胞催化反应的影响
Fig.3 The effect of reaction pH on whole-cell catalytic reaction
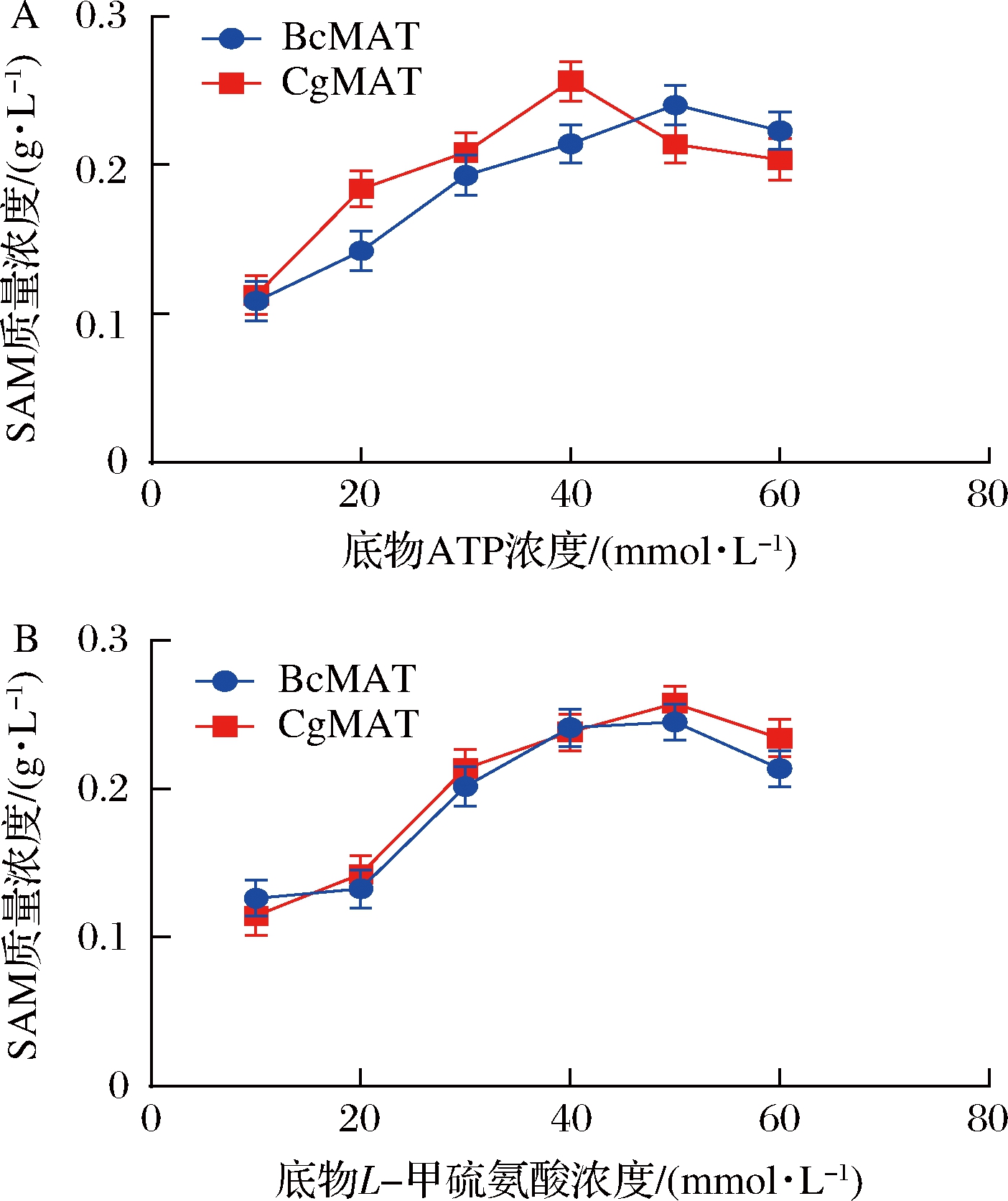
A-底物ATP浓度对全细胞催化的影响; B-底物L-甲硫氨酸浓度对全细胞催化的影响
图4 底物浓度对全细胞催化反应的影响
Fig.4 The effect of the concentration of the substrate on the catalytic reaction of whole cells
底物ATP浓度优化时,控制底物L-甲硫氨酸浓度为50 mmol/L,测定不同底物ATP浓度(10~60 mmol/L)下的SAM含量,以确定全细胞转化体系中ATP的最佳浓度。结果显示,E.coli BL21/pETDuet1-BcmetK重组菌株全细胞生物转化的最适ATP浓度为50 mmol/L,此时SAM含量为240 mg/L;E.coli BL21/pETDuet1-CgmetK重组菌株全细胞生物转化的最适ATP浓度为40 mmol/L,此时SAM含量为256 mg/L。
底物L-甲硫氨酸浓度的优化则是在上面优化的得到的最适ATP浓度下,测定不同浓度底物L-甲硫氨酸浓度(10~60 mmol/L)下SAM的含量,以确定全细胞转化体系中L-甲硫氨酸的最佳浓度。结果显示,E.coli BL21/pETDuet1-BcmetK重组菌株全细胞生物转化的最适L-甲硫氨酸浓度为50 mmol/L,此时SAM含量为245 mg/L;E.coli BL21/pETDuet1-CgmetK重组菌株全细胞生物转化的最适L-甲硫氨酸浓度为50 mmol/L,此时SAM含量为257 mg/L。
2.3.4 菌体浓度的优化
SAM全细胞转化过程中最佳菌体浓度是通过将OD600值约9.0的菌液按不同体积分数(0.2%~1.4%,间隔0.2%)加入到全细胞转化体系中进行转化合成,以确定最适的添加比例。20 mL反应液由50 mmol/L L-甲硫氨酸、适当浓度ATP (BcMAT:50 mmol/L、CgMAT:40 mmol/L)、50 mmol/L MgSO4、100 mmol/L K2SO4 组成,45 ℃、各自最适pH值(BcMAT:8.0,CgMAT:8.5)下反应5 h,HPLC检测SAM含量。结果如图5所示,当细胞添加比例为1.2% (体积分数)时,E.coli BL21/pETduet1-BcmetK重组菌株显示出最佳的生物转化活性,SAM含量为297 mg/L;而E.coli BL21/pETduet1-CgmetK重组菌株在细胞添加比例为0.8%(体积分数)时转化效果最佳,SAM含量达到307 mg/L。

图5 菌体浓度对全细胞催化反应的影响
Fig.5 The effect of cell concentration on the catalytic reaction of whole cells
2.4 对甲苯磺酸钠对全细胞催化合成SAM的影响
一些报告表明,大多数SAM合酶容易受到产物抑制,只有少数例外,特别是酿酒酵母的SAM合酶2不受产物抑制[20-22]。为了测试蜡状芽孢杆菌和谷氨酸棒杆菌来源的SAM合酶是否存在产物抑制,在含有50 mmol/L L-甲硫氨酸、适当浓度ATP (BcMAT:50 mmol/L、CgMAT:40 mmol/L)、50 mmol/L MgSO4、100 mmol/L K2SO4、适当菌体浓度[BcMAT:1.2% (体积分数)的OD600值为9.0的细胞悬浮液、CgMAT:0.8% (体积分数)的OD600值为9.0的细胞悬浮液]的20 mL反应体系中添加不同浓度(200~1 000 mmol/L)的pTSoNa,45 ℃、最适pH (BcMAT:8.0,CgMAT:8.5)下反应5 h后,加入6%(体积分数)HClO4终止反应。对照组则不添加pTSoNa。
结果如图6所示,E.coli BL21/pETduet1-BcmetK重组菌株全细胞转化过程pTSoNa的最优添加浓度为600 mmol/L,此时,SAM含量达到464 mg/L;E.coli BL21/pETduet1-CgmetK重组菌株全细胞转化过程pTSoNa的最优添加浓度为800 mmol/L,此时,SAM含量达到528 mg/L,由此可见,pTSoNa的添加可缓解产物SAM对SAM合酶的抑制作用,这也证实了先前文献报道的pTSoNa可改善SAM合酶的产物抑制现象的研究结论[23]。在800 mmol/L pTSoNa存在下,E.coli BL21/pETduet1-CgmetK重组菌株转化5 h 的SAM含量达到528 mg/L,相比对照提高了1.72倍(对照为不添加pTSoNa,SAM含量只能达到307 mg/L)。E.coli BL21/pETduet1-BcmetK重组菌株也表现出相似的特性,在600 mmol/L pTSoNa存在时,转化5 h,SAM含量达到464 mg/L,相比对照提高了1.56倍(对照组为不添加pTSoNa,SAM含量只能达到297 mg/L)。由此可见,E.coli BL21/pETduet1-CgmetK重组菌株合成SAM的能力优于E.coli BL21/pETduet1-BcmetK重组菌株,即谷氨酸棒杆菌来源的SAM合酶催化能力优于蜡状芽孢杆菌来源的,更有利于SAM的合成。
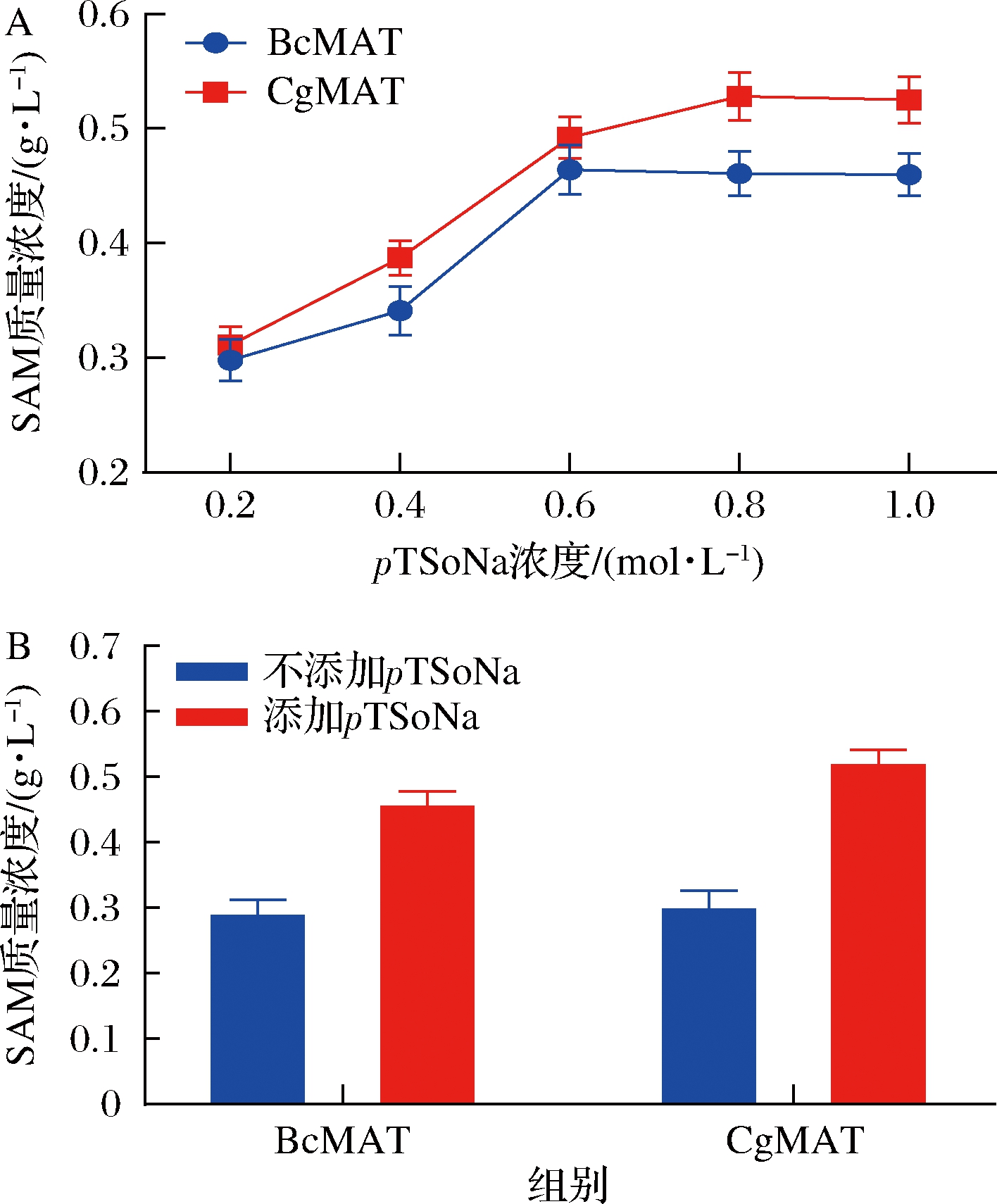
A-不同浓度对甲苯磺酸钠对全细胞催化合成SAM的影响; B-对甲苯磺酸钠对全细胞催化合成SAM的影响
图6 对甲苯磺酸钠对全细胞催化合成SAM的影响
Fig.6 The effect of sodium p-toluenesulfonate on the catalytic reaction of methionine adenosyltransferase
2.5 序列比对
为了更好地理解为什么在相似条件下两种重组酶的催化效率存在差异,将从Uniprot 数据库查询到的BcMAT(登录号:Q816Q8)和CgMAT(登录号:Q9K5E4)蛋白序列使用NCBI的COBALT工具进行比对分析。比对结果如图7所示,比对显示CgMAT比BcMAT的蛋白序列多8个氨基酸,这两种来源的SAM合酶具有超过50%的相似性,不同的残基可能位于具有催化作用的区域,所以这两种SAM合酶的催化能力存在差异。

A-图形展示;B-序列比对
Fig.7 CgMAT (登录号:Q9K5E4)和BcMAT (登录号:Q816Q8)的氨基酸序列比对
Fig.7 Comparison of amino acid sequences of CgMAT (accession number Q9K5E4) and BcMAT (accession number Q816Q8)
3 讨论
SAM在许多领域都有重要的应用,包括化学治疗和制药工业。浙江工商大学的YU等[25]通过在大肠杆菌中E.coli BL21(DE3)引入E.coli K-12 (W3110)来源的SAM合酶编码基因metK,重组菌株发酵培养条件优化,摇瓶发酵SAM含量可达128.2 mg/L,5 L发酵罐中发酵8 h后SAM的最高产量为300.9 mg/L,实现了SAM的发酵法合成。本研究在大肠杆菌E.coli BL21(DE3)中异源表达了蜡状芽孢杆菌和谷氨酸棒杆菌来源的SAM合酶,构建了重组大肠杆菌E.coli BL21/pETDuet1-BcmetK和E.coli BL21/pETDuet1-CgmetK,实现了SAM的一步法全细胞催化合成。并针对重组菌株全细胞生物转化合成SAM条件进行了优化,研究结果显示,重组大肠杆菌E.coli BL21/pETDuet1-BcmetK在最优条件[L-甲硫氨酸浓度50 mmol/L、ATP浓度50 mmol/L、1.2%(体积分数) OD600值约9.0的菌体浓度、600 mmol/L pTSoNa、50 mmol/L MgSO4、100 mmol/L K2SO4、45 ℃、pH 8.0]下,连续转化5 h,获得了464 mg/L的SAM。重组大肠杆菌E.coli BL21/pETDuet1-CgmetK在最优条件[L-甲硫氨酸浓度50 mmol/L、ATP浓度40 mmol/L、0.8%(体积分数) OD600值约9.0的菌体浓度、800 mmol/L pTSoNa、50 mmol/L MgSO4、100 mmol/L K2SO4、45 ℃、pH 8.5]下,连续转化5 h,获得了528 mg/L的SAM,可见,谷氨酸棒杆菌来源的SAM合酶催化能力优于蜡状芽孢杆菌来源的,更有利于SAM的合成。
该研究成功构建了SAM合成菌株,虽然产量水平不高,但为SAM的可持续生物合成提供了重要借鉴。接下来的研究中,为进一步改善SAM的产量,将从以下几个方面继续展开研究:(1)对SAM合酶进行挖掘,寻找高效酶;(2)针对SAM合酶的产物抑制现象进行理性改造,以缓解/解除反馈抑制;(3)尝试在不同宿主,如毕赤酵母、酿酒酵母、枯草芽孢杆菌、谷氨酸棒杆菌中进行SAM合成菌株的构建;(4)理性改造/酶表达体系优化提高SAM合酶的酶活力;(5)构建ATP再生系统,降低SAM合成成本[26]。
[1] MORRISONL D, SMITH D D, KISH S J.Brain S-adenosylmethionine levels are severely decreased in Alzheimer′s disease[J].Journal of Neurochemistry, 2010, 67(3):1 328-1 331.
[2] IZU H J, SHOBAYASHI M, MANABE Y, et al.Sake yeast suppresses acute alcohol-induced liver injury in mice[J].Bioscience, Biotechnology, and Biochemistry, 2006, 70(10):2 488-2 493.
[3] HOSEA BLEWETT H J.Exploring the mechanisms behind S-adenosylmethionine (SAMe) in the treatment of osteoarthritis[J].Critical Reviews in Food Science and Nutrition, 2008, 48(5):458-463.
[4] LIU W, TANG D D, SHI R, et al.Efficient production of S-adenosyl-L-methionine from DL-methionine in metabolic engineered Saccharomyces cerevisiae[J].Biotechnology Bioengineering, 2019, 116(12):3 312-3 323.
[5] 牛卫宁, 左晓佳, 王丽衡, 等.S-腺苷甲硫氨酸制备方法的研究进展[J].化学与生物工程, 2009,26(3):1-5.
NIU W N, ZUO X J, WANG L H, et al.Development for the preparation of S-adenosylmethionine [J].Chemistry & Bioengineering, 2009, 26(3):1-5.
[6] MATOS J R, RAUSHEL F M, WONG C H.S-adenosylmethionine:Studies on chemical and enzymatic synthesis[J].Biotechnology and Applied Biochemistry, 1987, 9(1):39-52.
[7] 刘沛溢, 董函竹, 谭天伟.补加前体L-蛋氨酸对高密度发酵生产S-腺苷-L-蛋氨酸的影响[J].生物工程学报, 2006, 22(2):268-272.
LIU P Y, DONG H Z, TAN T W.Effect of feeding pre-L-methionine on high-cell-density fermentation for S-adenosyl-L-methionine production [J].Chinese Journal of Biotechnology, 2006, 22(2):268-272.
[8] MARKHAM G D, PAJARES M A.Structure-function relationships in methionine adenosyltransferases[J].Cellular and Molecular Life Sciences, 2009, 66(4):636-648.
[9] KAMARTHAPU V, RAO K V, SRINIVAS P N B S, et al.Structural and kinetic properties of Bacillus subtilis S-adenosylmethionine synthetase expressed in Escherichia coli[J].BBA-Proteins and Proteomics, 2008, 1 784(12):1 949-1 958.
[10] SULLIVAND M, HOFFMAN J L.Fractionation and kinetic properties of rat liver and kidney methionine adenosyltransferase isozymes[J].Biochemistry, 1983, 22(7):1 636-1 641.
[11] KOTB M, KREDICH N M.S-Adenosylmethionine synthetase from human lymphocytes,purification and characterization[J].Journal of Biological Chemistry, 1985, 260(7):3 923-3 930.
[12] LU Z J, MARKHAM G D.Enzymatic properties of S-adenosylmethionine synthetase from the archaeon Methanococcus jannaschii[J].Journal of Biological Chemistry, 2002, 277(19):16 624-16 631.
[13] PORCELLI M, ILISSO C P, DE LEO E, et al.Biochemical characterization of a thermostable adenosylmethionine synthetase from the archaeon Pyrococcus furiosus with high catalytic power[J].Applied Biochemistry Biotechnology, 2015, 175(6):2 916-2 933.
[14] CHIANG P K, CANTONI G L.Activation of methionine for transmethylation,purification of the S-adenosylmethionine synthetase of bakers′ yeast and its separation into two forms[J].Journal of Biological Chemistry, 1977, 252(13):4 506-4 513.
[15] SHIOMI N, FUKUDA H, FUKUDA Y, et al.Production of S-adenosyl-D,L-homocysteine by Saccharomyces cerevisiae cells transformed with an ethionine resistance gene[J].Agricultural and Biological Chemistry, 1990, 54:1 595-1 596.
[16] SHIOMI N, FUKUDA H, MURATA K, et al.Improvement of S-adenosylmethionine production by integration of the ethionine-resistance gene into chromosomes of the yeast Saccharomyces cerevisiae[J].Applied Microbiology Biotechnology, 1995, 42(5):730-733.
[17] HE J, DENG J, ZHENG Y, et al.A synergistic effect on the production of S-adenosyl-L-methionine in Pichia pastoris by knocking in of S-adenosyl-L-methionine synthase and knocking out of cystathionine-beta synthase[J].Journal of Biotechnology, 2006, 126(4):519-527.
[18] QIN X L, LU J, ZHANG Y, et al.Engineering Pichia pastoris to improve S-adenosyl-L-methionine production using systems metabolic strategies[J].Biotechnology and Bioengineering, 2020, 117(5):1 436-1 445.
[19] MENANT A, BARBEY R, THOMAS D.Substrate-mediated remodeling of methionine transport by multiple ubiquitin-dependent mechanisms in yeast cells[J].The EMBO Journal, 2006, 25(19):4 436-4 447.
[20] CHEN Y W, TAN T W.Enhanced S-adenosylmethionine production by increasing ATP levels in baker′s yeast (Saccharomyces cerevisiae)[J].Journal of Agricultural and Food Chemistry, 2018, 66(20):5 200-5 209.
[21] CHOI E S, PARK B S, LEE S W, et al.Increased production of S-adenosyl-L-methionine using recombinant Saccharomyces cerevisiae sake K6[J].Korean Journal of Chemical Engineering, 2009, 26(1):156-159.
[22] CHU J, QIAN J C, ZHUANG Y P, et al.Progress in the research of S-adenosyl-L-methionine production[J].Applied Microbiology Biotechnology, 2013, 97(1):41-49.
[23] YIN C L, ZHENG T, CHANG X.Biosynthesis of S-adenosylmethionine by magnetically immobilized Escherichia coli cells highly expressing a methionine adenosyltransferase variant[J].Molecules, 2017, 22(8):1 365.
[24] LIN B X, TAO Y.Whole-cell biocatalysts by design[J].Microbial Cell Factories, 2017, 16:106.
[25] YU P, ZHU P Z.Improving the production of S-adenosyl-L-methionine in Escherichiacoli by overexpressing metK[J].Preparative Biochemistry and Biotechnology, 2017, 47(9):867-873.
[26] YAN G B, LI X, YANG J, et al.Cost-effective production of ATP and S-adenosylmethionine using engineered multidomain scaffold proteins[J].Biomolecules, 2021, 11(11):1 706.