微生物基因组精简是通过删减非必需基因,简化基因组组成,优化生化代谢网络,提高对底物和能量的利用效率,获得更适用的宿主菌株[1-2]。微生物基因组简化一般选用模式菌株,主要因其具有明晰的遗传背景和成熟的遗传学工具。近年来随着生物信息学技术的发展和分子操作平台的开发,非模式菌的基因组简化和底盘化改造受到重视[3-4]。
微生物基因组的大片段删除手段包括同源重组技术、CRISPR技术和转座重组技术[5]。在酿酒酵母和粟酒裂殖酵母中,通过线性DNA片段同源重组已获得了一系列基因组精简菌株[6-8]。CRISPR/Cas9介导的大片段删除技术是利用成对sgRNA识别不同的靶位点,引导Cas9产生DNA双链断裂,进而删除2个靶位点之间的序列[9],可作为基因组删减的工具。
产甘油假丝酵母(Candida glycerinogenes)具有多重抗逆、可高温浓醪发酵等优点[10-11],有成为强健工业生物宿主的潜质。前期实验室已完成全基因组测序和适合该菌的CRISPR/Cas9系统的构建[12-13]。基于其多种抗逆性,已成功改造应用于2-苯乙醇[14]和纤维素乙醇等的合成[15-16]。为使产甘油假丝酵母底盘化,本研究以大片段删除的方式对其基因组进行精简,在获得几个不同缺失程度的突变株后,基于耐受能力和发酵特性进行表征分析,以期为非模式菌底盘化相关研究提供借鉴。
1 材料与方法
1.1 菌株和质粒
本研究使用的菌株和质粒参见表1。
表1 本研究使用的菌株和质粒
Table 1 Strains and plasmids used in this study

菌株和质粒特征描述来源E.coli JM109克隆转化宿主实验室保藏C.glycerinogenes WTC.glycerinogenes野生型菌株实验室保藏C.glycerinogenes URA5C.glycerinogenes尿嘧啶营养缺陷型菌株,ura5基因缺失实验室保藏C.glycerinogenes U-/UC.glycerinogenesURA5衍生菌株,ura5基因整合表达本研究构建
续表1

菌株和质粒特征描述来源C.glycerinogenesΔ7.8 kbC.glycerinogenesURA5衍生菌株,删除7.8 kb序列本研究构建C.glycerinogenesΔ50 kbC.glycerinogenesURA5衍生菌株,删除50 kb序列本研究构建C.glycerinogenesΔ25 kbC.glycerinogenesURA5衍生菌株,删除25 kb序列本研究构建pMY-Cas9pMD19-T-Pgap-CgCas9-Taox1实验室保藏pMY-sgRNApMD19-T-Pgap- sgRNA -Taox1实验室保藏
1.2 引物设计
本研究所用引物参见表2。

续表2

引物名称序列(5'→3')Δ50 kb-Ri-F/RCCCAAGTCCCTCAAAAAACT/CTTGCTATTGAGTTTTTTTTTTATTTTTTATTTΔ25 kb-L1-sg-FAAATTTGGAAGCGTTAGCACGTTTTAGAGCTAGAAATAGCAAGTTΔ25 kb-L1-sg-RGTGCTAACGCTTCCAAATTTGACGAGCTTACTCGTTTCGΔ25 kb-L1-6 bp-F/RCTAGCAAATTTCTGATGAGTCCGTGAGGACGAAAC/ATCAGAAATTTGCTAGCAGAGCGGCCGΔ25 kb-L2-sg-FATTTGGAAGCGTTAGCACCGGTTTTAGAGCTAGAAATAGCAAGTTΔ25 kb-L2-sg-RCGGTGCTAACGCTTCCAAATGACGAGCTTACTCGTTTCGΔ25 kb-L2-6 bp-F/RCTAGCCCAAATCTGATGAGTCCGTGAGGACGAAAC/ATCAGATTTGGGCTAGCAGAGCGGCCGΔ25 kb-R1-sg-FTAAAGTTGTGTAACTGGCGAGTTTTAGAGCTAGAAATAGCAAGTTΔ25 kb-R1-sg-RTCGCCAGTTACACAACTTTA GACGAGCTTACTCGTTTCGΔ25 kbR1-6 bp-F/RCTAGCACTTTACTGATGAGTCCGTGAGGACGAAAC/ATCAGTAAAGTGCTAGCAGAGCGGCCGΔ25 kb-R2-sg-FGTTTGCACCTTATGAGTATC GTTTTAGAGCTAGAAATAGCAAGTTΔ25 kb-R2-sg-RGATACTCATAAGGTGCAAACGACGAGCTTACTCGTTTCGΔ25 kb-R2-6 bp-F/RCTAGCGCAAACCTGATGAGTCCGTGAGGACGAAAC/ATCAGGTTTGCGCTAGCAGAGCGGCCGΔ25 kb-1kbLe-FCAGAGTACAGAAGATTAAGTGAGACTAGTAGTAACCCTAATTACTGTCTTCGGAACTTΔ25 kb-1kbLe-RCTAATACCGTCGCCGTTTTCGAG GAATATCTAAATTTGATTCATTTCTCTGCGCTTCTTTTTTTΔ25 kb-1kbRi-FGAGTGCATTGGTACAATGGTGTTCAATTGGTTTCATTCGAAGCAAACTATAGTACAGGTΔ25 kb-1kbRi-RATCGCATTTCGCTGCGCTCTTCATCAATTTTTGTTTTATTAGTAAGAGGATT-AGGAATATCTTTATAAAΔ25 kb-OUT-F/RCTGTTGTCACGTGTTATACACTAGG/TCAGACTCTCCCAGAAAAAAAGTGAATΔ25 kb-IN-F/RCTCTGTAGAAAAATAACCTTATATTTGGCTAA/GGTTGTCAAGAAGATATCAGACAACΔ25 kb-Le-F/RTTTGTTTTACAACTTGATTTGAAATGAAATTTATC/CGTAGTGTTTCGAAGAAATAACATCΔ25 kb-Ri-F/RCCTTTCACGTACTGAATGG/AATCTGATAACGAAACTATGCTAACRT Hxt4-F/RTTGCCTTTGGTGGTTTCGTC/ACGAAATCGGGCATGTTGACRT GLK1-F/RAGCAGACGTCGTTGAATTCG/GCAAAACGGCAACAATGTCCRT PFK-α-F/RTGCGTTGGTTGTTTGTGGTG/AACCAATGAAGGCCACTCACRT PFK-β-F/RAATCAGCGAACGCCTCAAAG/ACCACCTTGTTGAACATGGCRT PYK1-F/RAGCGTGGAAAAGCTTGTTGC/TTGTCAATGACGGACTGGTGRT HOG1-F/RTGCTGACCTCCAGACCATTG/AGTCAATCGCATCAGGGTCGRT OLE1-F/RTTTACTCGGCTCGCAACCTT/TGTGAGCAGCGTTTGAATGCRT TPS1-F/RTTTGCGTGGTTCTTCAGCTC/ACCAACCAAATGCGTTTCCCRT RGT1-F/RCCACGCTCGAGGTCTAACTC/TGACTCTACCGAGCCTCTCCRT SKO1-F/RCAAAAGTCCCTGCCTCCCTT/TAGGCCCAGGAGTTAGACCCRT GPD1-F/RGTGGTGCCTTATCAGGTGCT/ATCTGTCCTGCCCATGTTGGRT FLO1-F/RACCACCAACTTAGAAGGCGG/TGGGTTTCAGTCGATGGTGGRT G6PD-F/RGGGCCAGCTCTCAGAAACAA/CACCACGTCCTTCAGTTCCART 6PGDH-F/RGGTGAATGGAACGAGGGTGT/TTCTCCAACCAGACTGTGCCRT ALD1-F/RAATCTTTGGCCCTGTTGTGG/AACTTGGCAGCCACAGAATGRT ADH1-F/RTGTTGCCAAGGGTGAAAACG/TTAGGGCAGTTTGGTTCAGC
注:下划线序列表示与目的扩增片段的同源臂序列,F表示正向引物,R表示反向引物
1.3 试剂与仪器
酵母膏、蛋白胨,Oxoid公司;实验用各种限制性内切酶、ExTaq DNA聚合酶,TaKaRa公司;质粒小量抽提试剂盒、胶回收试剂盒、柱回收试剂盒和酵母DNA提取试剂盒,生工生物工程(上海)股份有限公司;引物,上海亦欣生物科技有限公司。
P680高效液相色谱仪,美国戴安公司;ALS1296 PCR仪、PowerPac Basic琼脂糖凝胶电泳仪、Aminex HPX-87H液相色谱柱,美国Bio-Rad公司;AlphaImager凝胶成像仪,Alpha Innotech公司。
1.4 培养基与培养方法
LB培养基(g/L):酵母粉5.0,蛋白胨10.0,NaCl 10.0。氨苄青霉素抗性平板需额外添加100 μg/mL氨苄青霉素。
YPD培养基(g/L):酵母粉10.0,蛋白胨20.0,葡萄糖20.0。固体培养基加入琼脂20.0 g/L。
玉米浆种子培养基(g/L):葡萄糖100.0,尿素2.0,玉米浆8.0。
玉米浆发酵培养基(g/L):葡萄糖300.0,尿素2.0,玉米浆8.0。
马铃薯葡萄糖肉汤培养基(potato dextrose broth,PDB)培养条件:挑取单菌落接种于10 mL YPD培养基,30 ℃,200 r/min培养,12 h后转接50 mL YPD培养基培养,初始生物量控制为0.2,30 ℃,200 r/min培养48 h。
发酵条件:挑取单菌落接种于50 mL玉米浆种子培养基,于往复式摇床32 ℃、110 r/min培养,19 h后转接50 mL玉米浆发酵培养基培养,初始生物量控制为0.5,于往复式摇床32 ℃、110 r/min培养。
1.5 pYM-sg的构建
根据文献[12]和[13]设计C.glycerinogenes敲除靶向目的片段两端所用的sgRNA。具体来说,通过使用sgRNAcas9 V2.0.10信息学软件比对基因组序列分析潜在靶点,来设计sgRNA引导序列,通过sg-F/R和6 bp-F/R进行反向扩增模板质粒pMY-sgRNA,然后经测序验证成功获得所需质粒。
1.6 URA5回补片段的构建
质粒pMY-sgRNA上具有URA5基因表达框。
根据目的缺失片段设计带有同源序列的引物Donor Le-F/R和Donor Ri-F/R,以C.glycerinogenes WT基因组为模板,经PCR获得URA5回补片段左侧同源臂Donor Le和Donor Ri。将Donor Le与经限制性核酸内切酶Xho I线性化的pMY-sgRNA连接,这一步获得的质粒再经限制性核酸内切酶Cla I酶切线性化进一步与Donor Ri连接,即获得含有目的片段同源区域的URA5回补片段。以Donor Le-F和Donor Ri- R为上下游引物、以构建出的质粒为模板,经PCR可获得相应的URA5回补片段。
1.7 生物量测定
取1 mL发酵液适当稀释,使用分光光度计于600 nm下测定吸光值(OD600)。
1.8 发酵液中残糖、甘油和乙醇含量测定
离心(10 000×g,10 min)收集发酵上清液,用微孔水系滤膜(0.22 μm)过滤,最后利用HPLC测定浓度。具体条件设置参照嵇豪[17]的方法。以甘油含量(g/L)与生物量(OD600)的比值作为单位菌体甘油产量。
1.9 实时荧光定量分析
挑取C.glycerinogenes单菌落于玉米浆种子培养基,32 ℃、往复式摇床110 r/min条件下摇瓶培养,19 h后转接玉米浆发酵培养基,32 ℃、110 r/min培养24 h。总RNA提取参考JI等[18]的方法。RT-qPCR反应体系与条件参见南京诺唯赞试剂盒说明书。引物序列见表1。相对转录水平的计算和分析参考LIVAK 等[19]的方法。
1.10 胞内辅酶Ⅱ NADP(H)的测定
酵母细胞收集后用液氮淬灭,随后使用辅酶Ⅱ检测试剂盒对NADP(H)进行提取、测定,具体操作按苏州科铭试剂盒说明书进行。
2 结果与分析
2.1 成对sgRNA介导的大片段删除菌株构建
本研究将不含必需基因的长片段确定为敲除区域[20]。其中,7.8 kb长片段为2段互为回文序列9 537 bp IRL1(0~9 537 nt)和9 538 bp IRR2(22 173~31 711 nt)之间的非编码区序列(经NCBI BLAST后,未比对出基因,下同)(11 433~19 194 nt)(图1-a)。50 kb片段组成为:一对基因组上的长重复区域9 537 bp IRL1(10 905~20 442 nt)和9 538 bp IRR2(33 078~50 093 bp)、一个含未知蛋白的10 905 bp DNA序列(1 650 bp假设蛋白C5L36_0A06700)、长重复序列间的非编码区12 636 bp序列、另一个含未知蛋白的7 477 bp DNA序列(1 653 bp假设蛋白C5L36_0A06800)(图1-b)。25 kb长片段是指含有非编码区的1 296 bp DNA序列、一对长重复序列其中一支12.68 kb片段IRL2以及10 970 bp的非编码区序列(图1-c)。

a-7.8 kb长片段;b-50 kb片段;c-25 kb长片段
图1 本研究选择的敲除区域示意
Fig.1 Deletion areas selected in this study
以前期构建的CRISPR/Cas9瞬时表达系统[12-13]为基础,采用LiAc/SS carrier DNA/PEG的方法将敲除元件Cas9、若干成对的sgRNA和URA5回补片段转化出发菌株C.glycerinogenesURA5(图2)。

图2 成对sgRNA介导的大片段敲除示意图
Fig.2 Large fragment deletion guided by pairs of sgRNA
进行7.8 kb长片段敲除鉴定选用的引物如图3-a所示,PCR验证结果如图3-b所示。经类似过程,获得25、50 kb片段的敲除菌(图3)。
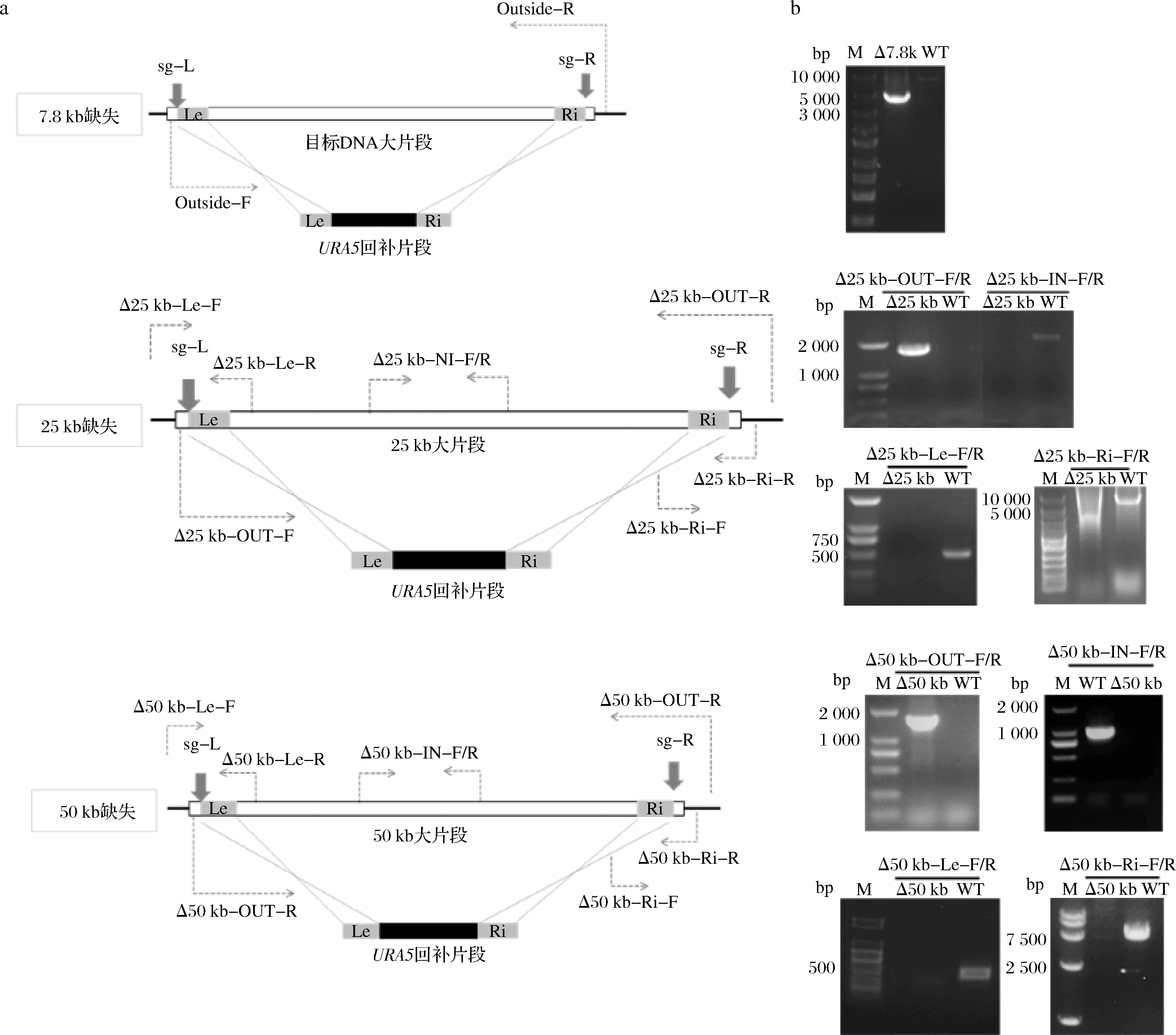
M-核酸分子标准;OUT-F/R-靶片段外部的PCR验证引物;IN-F/R-靶片段内部的PCR验证引物;
Le-F/R-靶片段上游的PCR验证引物;Ri-F/R-靶片段下游的PCR验证引物
a-验证缺失选用引物示意图;b-核酸电泳验证
图3 C.glycerinogenes片段缺失菌的构建过程
Fig.3 Construction of the genomic deletion C.glycerinogenes
2.2 缺失菌表型评价
表型分析发现,常规培养下Δ50 kb、Δ25 kb菌与野生菌的生物量相近,而Δ7.8 kb菌的最终生物量降低20%。42 ℃下,Δ50 kb菌的最终生物量相比对照无差异,Δ7.8 kb、Δ25 kb菌的生物量下降了20%左右(图4-a)。Δ25 kb菌缺失了一段基因组上重复序列,真核生物基因组上的重复序列可能参与高温胁迫下菌株的自我保护功能的发挥[21],因而造成Δ25 kb菌的耐高温性能减弱。在多种胁迫下,C.glycerinogenes的各缺失菌株仍持有耐受能力(图4-b),表明缺失的大片段DNA与上述胁迫下菌体抗逆能力无强关联性。甘油发酵中(图4-c),12 h时Δ7.8 kb菌的生物量较野生菌高出14.0%。在72 h时相较野生菌,各缺失菌生物量均降低20.0%左右。缺失菌稳定期生物量均低于对照,表明大片段缺失会导致最终生物量降低。发酵至108 h时,Δ7.8 kb菌残糖约为野生菌的62.5%,糖耗增加39.8 g/L,其他菌的残糖量则远高于对照菌,表明该7.8 kb片段缺失促进了糖耗。Δ7.8 kb菌甘油产量与野生菌相似,而单位菌体甘油产量达4.57,提高了22%,表明基因缺失促进了菌体的甘油合成能力。乙醇的合成是此发酵过程葡萄糖另一主要去向,野生菌和Δ7.8 kb菌乙醇产量分别为22.2和36.1 g/L,反映出Δ7.8 kb菌进入糖酵解途径的碳流有所增大,从而表现为糖耗增加。

a-在30和42 ℃的YPD摇瓶生长曲线;b-在胁迫条件下进行点滴平板实验;c-在高糖玉米浆发酵培养基中进行甘油发酵能力测定
图4 缺失菌表型
Fig.4 Characterization of genomic deletion strains
2.3 缺失菌表型差异分析
为了考察上述表型的变化与基因调控的潜在关系,以发酵初期生物量最大、单位菌体甘油产量最大的Δ7.8 kb菌为对象,考察葡萄糖代谢相关基因(HXT4、GLK1、PFK-α、PFK-β、PYK1)、高糖胁迫应答相关基因(HOG1、RGT1、SKO1)、抗逆相关基因(OLE1、TPS1、FLO1)、甘油合成关键基因(GPD1)、还原力相关基因(G6PD、6PGDH)、竞争途径乙醇合成关键基因(ALD1、ADH1)相对转录水平(图5)。相比野生菌株,Δ7.8 kb菌的G6PD上调了10倍,6PGDH上调了29倍,而其他基因转录变化并不明显,表明该菌的磷酸戊糖途径更为活跃,主代谢途径碳流可能受到了重新分配,使得菌株在发酵初期积累更高的生物量,同时能够提供更多的还原力,强化了菌株的甘油合成能力。为明确Δ7.8 kb菌的磷酸戊糖途径关键基因的过表达是否提高了该途径的通量,本研究进一步分析了与该途径密切相关的辅酶Ⅱ NADP(H)(表3),胞内NADPH水平明显提高,且NADPH/NADP+比值提高了100%,反映磷酸戊糖途径通量增加。C.glycerinogenes基因组片段的缺失可能对基因起到调控作用,改变了还原力水平,从而影响了生理特性。

图5 C.glycerinogenesΔ7.8 kb缺失菌甘油发酵过程中相关基因转录水平差异
Fig.5 Expression levels of genes related to glycerol ermentation in C.glycerinogenes Δ7.8 kb and WT
表3 野生型菌株与Δ7.8 kb菌胞内NADP(H)含量
Table 3 Contents of NADP(H) in C.glycerinogenes WT and Δ7.8 kb
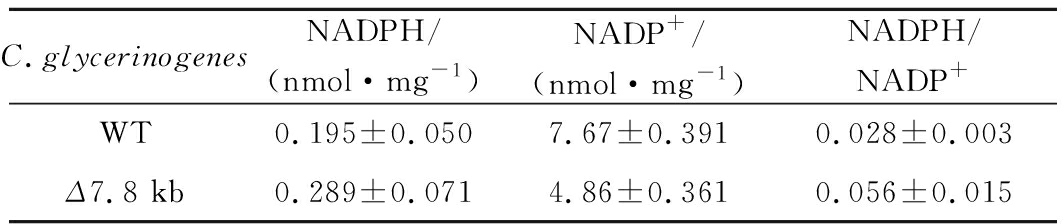
C.glycerinogenesNADPH/(nmol·mg-1)NADP+/(nmol·mg-1)NADPH/NADP+WT0.195±0.0507.67±0.3910.028±0.003Δ7.8 kb0.289±0.0714.86±0.3610.056±0.015
3 结论
C.glycerinogenes具有优良的抗逆性能和发酵性能,为了更好地适应合成生物学改造的需要,对其进行基因组精简化的底盘化策略。不同于以往仅应用于靶点敲除,本研究发掘的CRISPR/Cas9系统在C.glycerinogenes基因组上可以进行长达50 kb大片段敲除,为非模式菌底盘化提供可行思路。常规、高温条件下的培养中,大片段缺失的菌株出现菌体量积累能力减弱。在高糖、高盐、糠醛、乙醇的环境胁迫下,本研究中基因组不同程度缺失菌株的耐受性能仍然良好。甘油发酵中,缺失菌生物量均下降。而其中Δ7.8 kb菌则出现单位菌体甘油产量增加、糖耗加速,且磷酸戊糖途径活跃,表明该基因组大片段的缺失可能造成了戊糖磷酸途径和糖酵解途径碳流的重新分配,从而促进了菌体甘油合成效率。本研究表明CRISPR/Cas9系统可用于C.glycerinogenes大片段缺失、进行基因组简化研究,为后续非模式二倍体酵母的底盘化研究提供借鉴。后续对上述大片段基因组功能的研究能够进一步帮助理解该菌的甘油代谢合成调控机制。
[1] VICKERS C E, BLANK L M, KRÖMER J O.Grand challenge commentary:Chassis cells for industrial biochemical production[J].Nature Chemical Biology, 2010, 6(12):875-877.
[2] FOLEY P L, SHULER M L.Considerations for the design and construction of a synthetic platform cell for biotechnological applications[J].Biotechnology and Bioengineering, 2010, 105(1):26-36.
[3] BU Q T, YU P, WANG J, et al.Rational construction of genome-reduced and high-efficient industrial Streptomyces chassis based on multiple comparative genomic approaches[J].Microbial Cell Factories, 2019, 18(1):16.
[4] LIU J Q, ZHOU H B, YANG Z Y, et al.Rational construction of genome-reduced Burkholderiales chassis facilitates efficient heterologous production of natural products from proteobacteria[J].Nature Communications, 2021, 12(1):4347.
[5] GORYSHIN I Y, NAUMANN T A, APODACA J, et al.Chromosomal deletion formation system based on Tn5 double transposition:Use for making minimal genomes and essential gene analysis[J].Genome Research, 2003, 13(4):644-653.
[6] MURAKAMI K, TAO E, ITO Y, et al.Large scale deletions in the Saccharomyces cerevisiae genome create strains with altered regulation of carbon metabolism[J].Applied Microbiology and Biotechnology, 2007, 75(3):589-597.
[7] GIGA-HAMA Y, TOHDA H, TAKEGAWA K, et al.Schizosaccharomyces pombe minimum genome factory[J].Biotechnology and Applied Biochemistry, 2007, 46(Pt 3):147-155.
[8] RANG J, LI Y L, CAO L, et al.Deletion of a hybrid NRPS-T1PKS biosynthetic gene cluster via Latour gene knockout system in Saccharopolyspora pogona and its effect on butenyl-spinosyn biosynthesis and growth development[J].Microbial Biotechnology, 2021, 14(6):2 369-2 384.
[9] CANVER M C, BAUER D E, DASS A, et al.Characterization of genomic deletion efficiency mediated by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells[J].The Journal of Biological Chemistry, 2014, 289(31):21 312-21 324.
[10] JI H, ZHUGE B, ZONG H, et al.Role of CgHOG1 in stress responses and glycerol overproduction of Candida glycerinogenes[J].Current Microbiology, 2016, 73(6):827-833.
[11] YANG F, LU X Y, ZONG H, et al.Gene expression profiles of Candida glycerinogenes under combined heat and high-glucose stresses[J].Journal of Bioscience and Bioengineering, 2018, 126(4):464-469.
[12] ZHU M Y, SUN L, LU X Y, et al.Establishment of a transient CRISPR-Cas9 genome editing system in Candida glycerinogenes for co-production of ethanol and xylonic acid[J].Journal of Bioscience and Bioengineering, 2019, 128(3):283-289.
[13] 朱敏阳. 产甘油假丝酵母CRISPR-Cas9基因编辑系统的构建[D].无锡:江南大学, 2019.
ZHU M Y.Establishment of CRISPR-Cas9 genome editing system in Candida glycerinogenes[D].Wuxi:Jiangnan University, 2019.
[14] 王玉芹. 基于Candida glycerinogenes耐受性的2-苯乙醇强化合成研究[D].无锡:江南大学, 2021.
WANG Y Q.Genetic engineering improves 2-phenylethanol production in Candida glycerinogenes base on its tolerance mechanism[D].Wuxi:Jiangnan University, 2021.
[15] ZHAO M L, SHI D C, LU X Y, et al.Ethanol fermentation from non-detoxified lignocellulose hydrolysate by a multi-stress tolerant yeast Candida glycerinogenes mutant[J].Bioresource Technology, 2019, 273:634-640.
[16] 赵美琳. Candida glycerinogenes高浓度糠醛耐受机制及纤维素乙醇发酵应用[D].无锡:江南大学, 2019.
ZHAO M L.High concentration furfural tolerance mechanism of Candida glycerinogenes and application of cellulose ethanol fermentation[D].Wuxi:Jiangnan University, 2019.
[17] 嵇豪. Candida glycerinogenes HOG应答途径及渗透压耐受机制研究[D].无锡:江南大学, 2018.
JI H.HOG response pathway and the osmotic stress tolerance of Candida glycerinogenes[D].Wuxi:Jiangnan University, 2018.
[18] JI H, LU X Y, WANG C Y, et al.Identification of a novel HOG1 homologue from an industrial glycerol producer Candida glycerinogenes[J].Current Microbiology, 2014, 69(6):909-914.
[19] LIVAK K J, SCHMITTGEN T D.Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J].Methods, 2001, 25(4):402-408.
[20] 王建莉, 王小元.微生物基因组精简优化的研究进展[J].生物工程学报, 2013, 29(8):1 044-1 063.
WANG J L, WANG X Y.Advances in microbial genome reduction and modification[J].Chinese Journal of Biotechnology, 2013, 29(8):1 044-1 063.
[21] GILCHRIST C, STELKENS R.Aneuploidy in yeast:Segregation error or adaptation mechanism?[J].Yeast, 2019:3427.