酸面团发酵技术是一种具有悠久历史的食品加工技术,广泛应用于面包、馒头等发酵面制品中。酸面团是非常复杂的微生物生态系统,其中乳酸菌和酵母菌为优势微生物,它们通过自身的生长代谢以及相互作用产生大量的代谢产物,提高了酸面团产品的品质。除此之外还可能存在一些数量极少的其他菌(如霉菌、醋酸菌等)对酸面团产品的品质不会产生显著影响[1-2]。在酸面团的发酵过程中乳酸菌和酵母菌可以稳定共存,其中乳酸菌可以产生大量的乳酸、乙酸等有机酸,有机酸的产生和积累提高了面团的酸度,有利于改善面团的面筋结构,提高面团的延展性;酵母菌可以产生乙醇和CO2,增加面团的体积,赋予面团良好的结构特性[3]。2种微生物共同作用可以激活面团中内源酶的活性,促进大分子物质的分解,并产生大量的挥发性风味物质,例如游离氨基酸、苯乳酸、2-甲基丁酸和戊酸等重要风味活性成分,对发酵面制品的质构特性和感官特性均会产生积极作用[4-5]。此外,酸面团在微生物的作用下还会产生抗真菌化合物、胞外多糖和功能性肽等物质,对发酵面制品的保存期限、质构特性和营养特性等也具有积极影响[6-8]。
乳酸菌和酵母菌的种类和生长活性受多种因素的影响,DE VUYST等[9]对酸面团中乳酸菌和酵母菌进行了介绍,表明成熟酸面团中微生物的种类与酸面团的工艺参数密切相关,例如面粉类型、工艺参数等。MINERVINI等[10]的研究则表明酸面团中的微生物组成受内源性因素和外源性因素的影响,内源性因素主要由面粉的化学组成和微生物组成决定,外源性因素主要由温度和氧化还原电位决定,同时也阐述了酸面团中乳酸菌和酵母菌的相互作用。一般来说乳酸菌和酵母菌的最佳比例为100∶1,在该比例下乳酸菌和酵母菌具有良好的相互作用[11]。以往的文章大多侧重于酸面团中乳酸菌的种类和生长活性[12-13],对酸面团中酵母菌的种类以及酵母菌与乳酸菌的相互作用关注较少,而乳酸菌和酵母菌之间的相互作用对酸面团产品的品质至关重要,因此,本文不仅对酸面团的优势微生物组成进行了概述,还探讨了优势微生物(乳酸菌和酵母菌)之间的相互作用对酸面团产品品质的影响,以期为今后酸面团的研究提供理论依据。
1 酸面团的分类
如图1所示,目前根据酸面团的制备工艺可将其分为Ⅰ型酸面团、Ⅱ型酸面团和Ⅲ型酸面团[14-16]。Ⅰ型酸面团为传统的自然发酵酸面团,不需要额外添加菌种,以连续传代、连续发酵的方式生产,发酵温度一般低于30 ℃,发酵时间较长,同时需要定期向原酸面团中添加新的发酵底物和水,以维持微生物活性[17]。该类酸面团的微生物来自于自然环境,因此其组成比较复杂,其中的微生物代谢活性高,能产生多种生物活性成分,赋予发酵面制品更好的风味和营养价值[18]。但在发酵过程中Ⅰ型酸面团中的微生物组成极易受到各种内外因素的影响,例如面粉类型、发酵温度等[19],同时Ⅰ型酸面团也具有发酵时间长和微生物组成不可控制的劣势。Ⅱ型酸面团为工业化生产酸面团,通过人工接种目的菌株发酵酸面团,发酵温度高于30 ℃,可以显著地缩短发酵时间。该类酸面团中的微生物主要来自于人工接种,同时面粉本身也存在微生物,酸化速率快,生产效率高且易于控制,一般呈流体状态,选用的乳酸菌通常基于它们快速酸化的能力或产生特定风味成分的能力[9]。Ⅱ型酸面团中微生物代谢活性一般较Ⅰ型酸面团低。对Ⅱ型酸面团的研究通常集中在形成风味成分、抗真菌化合物,改善质地特性、营养特性等方面[20-23]。Ⅲ型酸面团是通过工业化生产得到的商业发酵剂,在Ⅱ型酸面团的基础上经过喷雾干燥、冷冻干燥等方法除去水分而得到粉末状固体,在干燥的过程中,乳酸菌的活力会受到影响,因而选用的菌株往往具有耐高温耐干燥的特性。该类酸面团在储存、运输等方面具有很大的优势,但风味成分有所损失[24]。3种类型酸面团的质量与酸面团中的微生物组成密切相关,通过研究酸面团中优势微生物组成及其相互作用,有利于了解酸面团发酵技术的独特优势,从而促进酸面团产品的发展。
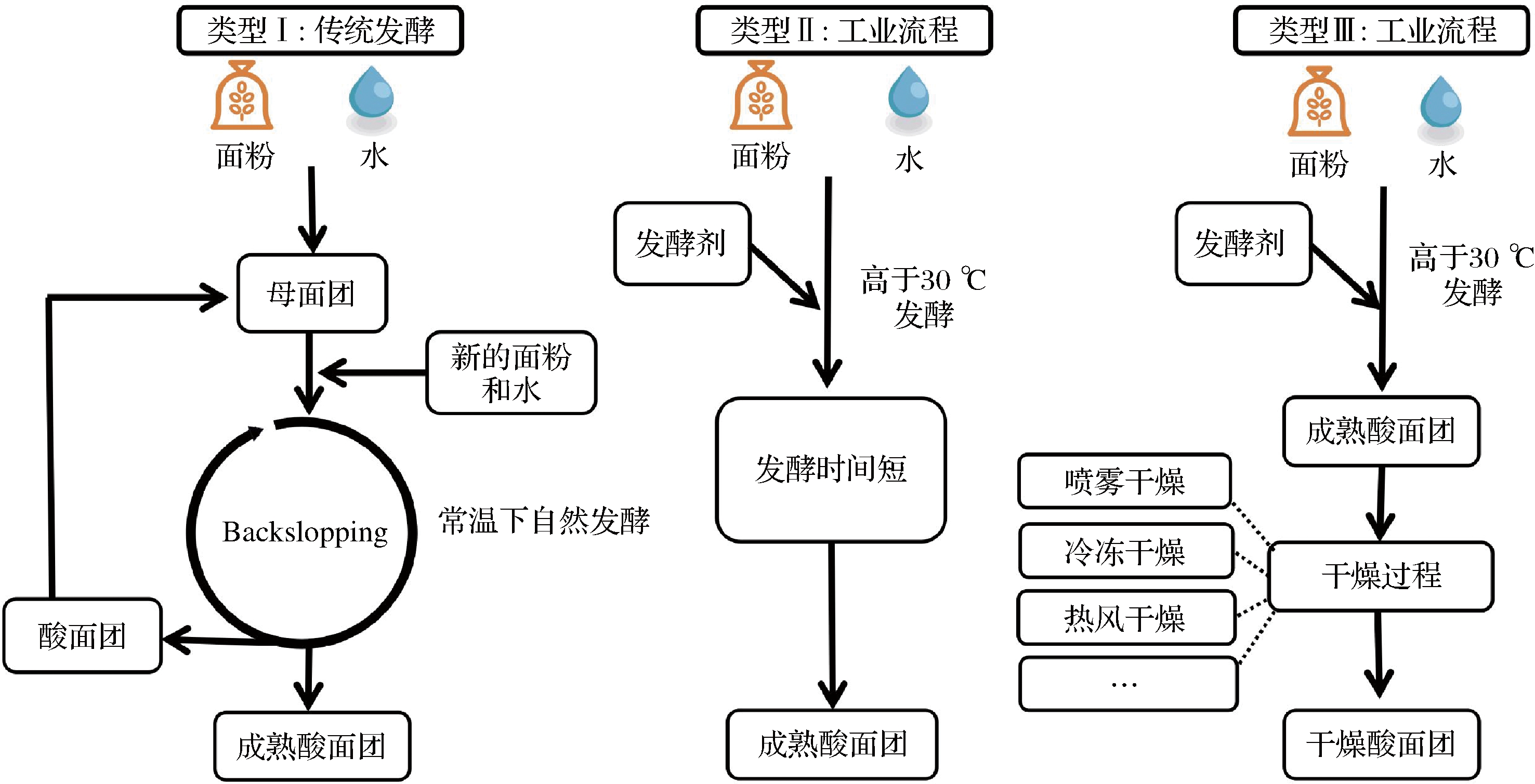
图1 酸面团的分类
Fig.1 Classification of sourdough
2 酸面团的微生物组成
酸面团中微生物组成的研究方法主要有培养法和非培养法,培养法是将酸面团中的微生物分离纯化后再结合菌落形态、生理生化特性、扩增子测序等完成菌种的鉴定,非培养法则是利用变性梯度凝胶电泳、时间温度梯度凝胶电泳或高通量测序技术对酸面团中的微生物组成进行检测[25]。一般传统的分离培养方法仅能分离纯化酸面团中的一部分微生物,无法代表酸面团中整个微生物群,变性梯度凝胶电泳则不能检测到种群数量占群落总数1%以下的菌落,而高通量测序技术可以检测出不可培养和低丰度微生物,因此常常被用到酸面团的微生物组成分析中,常用的高通量测序技术有扩增子测序、宏基因组测序和宏转录组测序3种[26-27]。此外,基于PCR的分类和识别技术,例如随机扩增多态性DNA PCR、细菌基因组重复序列PCR、单重或多重PCR和多位点序列分析技术,也已应用到酸面团中微生物的快速检测[28]。
2.1 酸面团发酵过程中的微生物动态变化
传统自然发酵酸面团的发酵初期,酸面团中的微生物组成主要取决于面粉中本身存在的微生物种群,其中包括乳酸菌、革兰氏阳性菌(例如芽胞杆菌)和革兰氏阴性菌(例如假单胞菌)、肠杆菌科、酵母菌等,每个微生物群的细胞数量通常不超过5.0 lg CFU/g[29]。面粉的水分活度较低,其中的微生物处于休眠状态,无法进行生长代谢,而当面粉与水混合后,面粉的水分活度增加,面团的氧化还原电位降低,有利于兼性厌氧菌(肠杆菌科和酵母菌)和乳酸菌的生长[30]。同时水分活度的增加激活了面粉中内源酶的活性,使其中的多糖分解为可发酵碳水化合物,酸面团中的乳酸菌利用可发酵碳水化合物产生乳酸和乙酸,导致酸面团的pH值不断降低,从而在后续的发酵过程中抑制了肠杆菌科的生长[31]。酸面团中的酵母菌对低pH值环境的耐受性较好,因此在连续发酵的过程中,乳酸菌和酵母菌对酸面团中的酸性环境适应性较强,逐渐成为其中的优势菌群,乳酸菌的数量从6.0 lg CFU/g增加到9.0 lg CFU/g,酵母菌的数量从5.0 lg CFU/g增加到8.0 lg CFU/g[32]。有研究发现奇亚籽酸面团在发酵过程中乳酸菌的数量从3.99 lg CFU/g 增加到9.25 lg CFU/g,肠杆菌科从初始的4.05 lg CFU/g在第10天降低到2.0 lg CFU/g以下,霉菌从发酵的第4天开始已无法检测出[33]。OSHIRO等[34]利用16S rRNA测序和目标化学物质的定量对连续发酵酸面团中微生物群落动态变化进行了研究,发现在60 d的连续发酵过程中乳酸菌和酵母菌逐渐主导酸面团的发酵,经过发酵后乳酸菌和酵母菌的数量分别达到了9.6 lg CFU/g和7.6 lg CFU/g。表明在酸面团发酵过程中,乳酸菌和酵母菌会逐渐成为其中的优势菌群,而肠杆菌科、假单胞菌和霉菌等会在发酵过程中逐渐减少至无法检测。
在酸面团中乳酸菌通常占主导地位,因此乳酸菌的生长与酸面团的品质密切相关,研究发现在酸面团的发酵过程中乳酸菌会经历相似的动态演变过程[35-36]:以肠球菌属、乳球菌属和明串珠菌属为优势菌;经过发酵后酸面团中特殊的乳酸菌逐渐增加,以乳杆菌属、片球菌属和魏氏菌属为主导;发酵完成后酸面团中以适应性专性异型发酵乳酸菌为主导,例如旧金山乳杆菌、发酵乳杆菌、植物乳杆菌等乳杆菌属。这种演变过程与乳酸菌对不同酸性条件的耐受性、碳水化合物代谢以及蛋白质代谢机制密切相关:首先,适应性异型发酵乳酸菌的碳水化合物代谢高度适应面团中的能量来源,通过麦芽糖磷酸酶以及以果糖为共同底物的戊糖磷酸途径利用麦芽糖,比同型发酵乳酸菌降解麦芽糖产生了更多的能量;其次,专性异型发酵乳酸菌(例如旧金山乳杆菌)生长代谢的适宜温度和pH值与酸面团发酵过程中温度和pH值的变化相匹配,因此更利于它们的生长代谢;最后,专性异型发酵乳酸菌具有多种应激反应机制来克服酸性环境、高温或低温环境、高渗透压或脱水环境以及缺乏可利用底物环境等劣势条件[37-39]。一般而言,Ⅰ型酸面团具有较低的发酵温度,发酵完成后其中以短乳杆菌、植物乳杆菌、旧金山乳杆菌等适应低温发酵的乳杆菌为主导;工业化的Ⅱ型酸面团与Ⅰ型酸面团相比具有更高的温度及含水量,因此发酵完成后其中以发酵乳杆菌、罗伊氏乳杆菌等耐热、耐酸的乳杆菌为主导[28]。
2.2 成熟酸面团中微生物组成
如表1所示,不同地区酸面团的微生物组成有所不同,总体而言成熟酸面团中的微生物主要为乳酸菌和酵母菌,其中乳酸菌物种的数量普遍高于酵母菌。目前酸面团中乳酸菌已鉴定出至少90多种,并以异型发酵乳杆菌为主要种群[40-41]。虽然已从酸面团中分离出明串珠菌属、魏氏菌属、片球菌属、乳球菌属、肠球菌属和链球菌属,但乳杆菌属是酸面团中最常见的细菌,其中有旧金山乳杆菌(Lactobacillus sanfranciscensis)、植物乳杆菌(Lactobacillus plantarum)、短乳杆菌(Lactobacillus brevis)、发酵乳杆菌(Lactobacillus fermentum)等[42-44]。酸面团中酵母菌的种类比乳酸菌要少,从酸面团中分离到的酵母菌有40多种,含酵母属、哈萨克斯坦属、维克多菌属、圆孢菌属和毕赤酵母属,其中最典型的酵母是酿酒酵母(Saccharomyces cerevisiae)、米勒念珠菌(Candida milleri)和矮小假丝酵母(Candida humilis),其他还分离到少孢酵母(Saccharomyces exiguus)、异常毕赤酵母(Pichia anomala)、汉逊德巴利酵母(Debaryomyces hansenii)等[17, 45]。
表1 不同地区成熟酸面团中的代表性菌株
Table 1 Representative strains in mature sourdough in different regions

地区鉴定方法主要乳酸菌主要酵母菌参考文献伊朗采用选择性琼脂培养进行菌株分离,DNA 扩增和16S rRNA、ITS区域测序分析Pediococcus pentosaceus, Weissella confusa, En-terococcus faecium, Leuconostoc citreumKluyveromyces marxianus, Kluyvero-myces lactis, Kluyveromyces aestuarii[46]爱尔兰采用选择性琼脂培养进行菌株分离,DNA 扩增和16S rRNA、25S rRNA测序分析Lactobacillus brevis, Lactobacillus acidophilus, Lactobacillus buchneri, Lactobacillus sanfran-ciscensis, Lactobacillus frumentiCandida glabrata, Saccharomyces cer-evisiae[47]法国采用Illumina 平台对16S rRNA 进行高通量测序Lactobacillus sanfranciscensis, Lactobacillus bre-vis, Lactobacillus curvatus, Lactobacillus xiang-fangensis, Lactobacillus heilongjiangensis, Lac-tobacillus koreensis, Pediococcus pentosaceus-[48]
续表1

地区鉴定方法主要乳酸菌主要酵母菌参考文献立陶宛采用选择性琼脂培养进行菌株分离,DNA 扩增和16S rRNA测序分析Leuconostoc mesenteroides, Lactobacillus planta-rum, Enteroccocus pseudoavium, Lactobacillus casei, Lactobacillus curvatusLactobacillus farrag-inis, Pediococcus pentosaceus-[49]土耳其采用选择性琼脂培养进行菌株分离,DNA 扩增和16S rRNA测序分析Lactobacillus brevis, Lactobacillus plantarum, Lactobacillus fermentum, Lactobacillus musae, Lactobacillus paralimentarius, Lactobacillus cur-vatus, Leuconostoc mesenteroides-[44]阿根廷采用Illumina 平台对16S rRNA 进行高通量测序Lactobacillus brevis, Lactobacillus mucosae, Lac-tobacillus plantarum, Lactobacillus sakei, Lacto-bacillus rhamnosus, Weissella cibaria-[31]奥地利采用MALDI-TOF MS对微生物蛋白质组学进行鉴定,采用Eurofins Genomics或Microsynth技术对16S rRNA、ITS区域测序分析Lactobacillus sanfranciscensis, Lactobacillus cur-vatus, Pediococcus pentosaceus, Lactobacillus paracasei、Lactobacillus diolivoransSaccharomyces cerevisiae, Kazach-stania humilis[50]意大利采用选择性琼脂培养进行菌株分离,DNA 扩增和16S rRNA、ITS区域测序分析Lactobacillus sanfranciscensis, Leuconostoc citre-um, Lactobacillus acidifarinae, Pediococcus pen-tosaceus, Lactobacillus curvatusKazachstania humilis, Saccharomyces cerevisiae, Kazachstania exigua[51]意大利采用焦磷酸测序技术对16S rRNA进行测序分析Pediococccus pentosaceus,Weissella cibaria,Lac-tococcus lactis, Lactobacillus sakei,Leuconostoc mesenteroides-[52]新西兰采用Illumina平台对16S rRNA和ITS区域进行高通量测序Lactobacillus fermentum, Lactobacillus planta-rum,Saccharomyces cerevisiae[53]比利时采用选择性琼脂培养进行菌株分离,DNA 扩增和16S rRNA和ITS区域测序分析Lactobacillus brevis, Lactobacillus fermentum, Lactobacillus plantarum, Leuconostoc citreum, Leuconostoc mesenteroides, Leuconostoc pseudom-esenteroidesSaccharomyces cerevisiae[54]日本采用选择性琼脂培养进行菌株分离,DNA 扩增和16S rRNA和LSU-rDNA测序分析Lactobacillus sakei, Lactobacillus brevis, Lacto-bacillus sanfranciscensis, Lactobacillus alimenta-rius, Lactobacillus pentosus, Lactobacillus vacci-nostercus-[55]中国采用焦磷酸测序技术对16S rRNA进行测序分析Lactobacillus sanfranciscensis, Lactobacillus paralimentarius, Lactobacillus crustorum, Lacto-bacillus plantarum, Weissella confusa, Lactoba-cillus fermentum-[56]中国采用选择性琼脂培养进行菌株分离,DNA 扩增和16S rRNA和28S RNA测序分析 Lactobacillus brevis, Lactobacillus sanfranciscen-sis, Weissella cibaria, Lactobacillus fermentum, Lactobacillus plantarum, Lactobacillus pontis, Lactobacillus paralimentariusSaccharomyces cerevisiae,Kazachstania humilis, Wickerhamomyces anomalus[57]中国采用Illumina技术对16S rRNA和 ITS区域进行高通量测序Lactobacillus sanfranciscensis, Lactobacillus pon-tisSaccharomyces cerevisiae,Kazachstania humilis[58]
3 乳酸菌与酵母菌的相互作用
3.1 协同作用
成熟酸面团中乳酸菌和酵母菌可以稳定共存,同时在酸面团发酵过程中乳酸菌和酵母菌之间还存在协同作用,产生这种相互作用的原因主要是基于这2种菌对碳水化合物、氨基酸等的代谢能力不同[59]。在乳酸菌和酵母菌的共发酵体系中,乳酸菌代谢产生有机酸,降低面团的pH值,在酸性环境下酵母菌开始酒精发酵,同时乳酸菌还会促进酸面团的蛋白水解,所产生的丙酮酸、二羧酸、氨基酸和脂肪酸等代谢产物可以促进酵母菌的生长[60]。当有机氮源和无机氮源同时存在时,酵母优先利用后者(如氨),而乳酸菌更偏好游离氨基酸,尤其是小肽。在生长过程中或由于酵母的自溶作用,酵母细胞可以分泌乳酸菌需要的维生素、氨基酸(例如脯氨酸、缬氨酸、异亮氨酸和甘氨酸等)和多肽,这些物质可作为生长因子促进乳酸菌的增长[61]。LIM等[62]发现在酸性条件下酿酒酵母可以提高鼠李糖乳杆菌的生存能力和活性,并且适当高的酵母菌浓度对鼠李糖乳杆菌的活性具有增强效果。SIEUWERTS等[63]在培养基中添加特定碳源以模拟酸面团发酵环境,发现在富含葡萄糖、果糖和乳糖的条件下,酿酒酵母与植物乳杆菌之间具有强烈的相互促进作用,而在富含半乳糖、麦芽糖、蔗糖和淀粉的条件下,两者的相互促进作用不明显,这主要与酿酒酵母和植物乳杆菌对碳源的代谢能力相关。
酸面团中乳酸菌和酵母菌的协同作用对酸面团产品的品质能产生积极影响,乌云等[64]研究发现酿酒酵母能与短乳杆菌、弯曲乳杆菌和植物乳杆菌稳定共生,通过混菌发酵可以缩短面团的发酵时间,并形成积极的发酵风味。WINTERS等[65]利用单一酵母发酵面团以及酵母和乳酸菌混菌发酵面团,比较了2种面团在风味物质、面包屑结构及感官评定上的差异,相较于单一酵母发酵面团,混菌发酵所得面包中苯甲醛、1-辛烯-3-醇、1-戊醇、1,2,3,4-四甲基苯和2-戊基呋喃等风味物质含量显著提高,且对面包的质构和感官品质都有积极影响。在乳酸菌和酵母菌的协同作用下还有利于酸面团中有益代谢物的产生,例如在乳酸菌和酵母菌的共发酵体系下,酿酒酵母可以将阿魏酸代谢为脱羧产物4-乙烯基愈创木酚,乳酸菌可以将阿魏酸代谢为二氢阿魏酸和4-乙烯基愈创木酚,而其中的乳酸菌在单一发酵中可能不会产生这些代谢产物,BOUDAOUD等[66]研究发现乳酸菌和酵母菌共发酵导致细胞外条件的改变,从而诱导了乳酸菌的脱羧酶和还原酶的活性,促进了二氢阿魏酸和4-乙烯基愈创木酚的产生,且这些代谢产物可以改善酸面团面包的营养和感官质量。
3.2 拮抗作用
在乳酸菌和酵母菌的共发酵体系中,少数乳酸菌和酵母菌之间还会存在拮抗作用。溶脂酵母代谢生成脂肪酸,会对乳酸菌的生长产生影响,而乳酸菌的代谢产物乳酸、4-羟基-苯乳酸、环肽则会抑制酵母菌的生长[67]。一些异型发酵乳酸菌经发酵后会产生乙酸,在酸面团的发酵后期(pH值约为4.0),大部分的乙酸未解离,因此可能会穿过细胞质进入细胞,会对一些酵母(例如酿酒酵母)的生长产生抑制作用[68]。在酸面团发酵过程中,乳酸菌和酵母菌也会同时快速消耗底物中的可溶性碳水化合物,只有在含有大量可溶性碳水化合物的条件下,微生物之间才会具有协同生长的作用。麦芽糖是麦芽糖阳性乳酸菌(例如旧金山乳杆菌)的首选碳源,该类乳酸菌可以通过麦芽糖磷酸化酶水解麦芽糖以释放葡萄糖-1-磷酸和葡萄糖,之后通过6-磷酸葡萄糖酸/磷酸酮醇酶途径代谢。这时面团中麦芽糖阳性酵母(例如酿酒酵母)的存在会与其竞争消耗麦芽糖,因此有可能会降低该类乳酸菌的代谢[69]。但一般情况下麦芽糖会在面粉中淀粉酶的存在下持续产生,在发酵过程中不会被耗尽,因此麦芽糖阳性乳酸菌和酵母菌也能实现稳定共存。而麦芽糖阴性酵母(例如假丝酵母和少孢哈萨克斯坦酵母)与旧金山乳杆菌建立了良好的营养互惠的关系。在共发酵过程中,麦芽糖阴性酵母能够将一些面粉中的低聚糖分解为果糖,为利用果糖作为电子受体的乳酸菌提供了关键优势,减少了乳酸菌和酵母菌对碳水化合物的竞争[31]。旧金山乳杆菌和假丝酵母之间的共生相互作用在温度为25~30 ℃、pH值为4.2~6.7的范围内最佳,而一般旧金山乳杆菌的最佳生长温度在32 ℃,假丝酵母的最佳生长温度为27 ℃[70]。
酸面团中乳酸菌和酵母菌的拮抗作用仅发生在少数菌种之间,且酸面团中的可溶性碳水化合物通常不会被耗尽,因此成熟酸面团中乳酸菌与酵母菌仍能达到稳定共存。YANG等[71]发现与单一发酵相比,混合发酵中的酿酒酵母和旧金山乳杆菌的最终细胞数量没有显著变化,表明酸面团原料为其提供了充足的营养成分,两者可以稳定共存,但在混合发酵中酿酒酵母的代谢仍受到了负面影响,旧金山乳杆菌的存在抑制了酸面团中与酿酒酵母相关风味物质(乙醇、2,3-戊二酮、3-甲基-1-丁醇、苯乙醇和2-甲基-1-丙醇)的产生。VOGELMANN等[72]发现将多种不同的乳酸菌和酿酒酵母共同接种于大米酸面团中连续发酵10 d,其中瑞士乳杆菌、桥乳杆菌的生长受到了抑制,占比减小,而植物乳杆菌、发酵乳杆菌的生长并没有受到影响,表明瑞士乳杆菌和桥乳杆菌与酿酒酵母之间存在拮抗作用。同时该学者发现这种拮抗现象只发生在底物基质有限的情况下,在底物充足时这些乳酸菌的生长通常不会被酿酒酵母抑制,因此对酸面团产品的品质不会产生明显的负面影响。
4 结论与展望
酸面团中的微生物具有丰富的多样性,乳酸菌和酵母菌是酸面团中的优势菌,微生物产生的有机酸、短肽、氨基酸、醇类和酯类等物质对酸面团的质地特性、营养特性以及感官特性均会产生积极影响。本文综述了酸面团发酵过程中微生物的组成变化,酸面团中优势菌群之间的相互作用对酸面团产品品质的影响,为酸面团的发展提供了理论基础。酸面团中除了乳酸菌和酵母菌起着重要作用外,近期的研究表明醋酸菌对酸面团产品的品质也具有积极影响[73],LI等[74]发现在酸面团中接种醋酸菌、乳酸菌和酵母菌的混合物可以使酸面团具有更好的发酵能力以及更好的氨基酸生物合成作用。醋酸菌在酸面团中可直接利用酵母菌的代谢产物乙醇作为碳源生成大量乙酸,乙酸的积累也可以促进酸面团中酵母菌的生长代谢,因此醋酸菌和酵母菌之间存在协同作用,但当乙酸过量时,酵母菌的活性又会受到抑制[75]。醋酸菌代谢还会产生草酸、酒石酸、葡萄糖酸等产物,对酸面团产品的风味具有积极影响[76]。在酸面团中醋酸菌的相关研究十分缺乏,关于醋酸菌和乳酸菌之间的相互作用还未见报道,随着宏基因组学、宏转录组学、蛋白质组学、代谢组学等的发展和应用,在酸面团中会发现更多更复杂的微生物组成及其相互的代谢活动,同时通过了解酸面团中微生物的动力学机制,有利于促进酸面团在发酵面制品中的应用。
[1] RIPARIV, G NZLE M G, BERARDI E.Evolution of sourdough microbiota in spontaneous sourdoughs started with different plant materials[J].International Journal of Food Microbiology, 2016, 232(6):35-42.
NZLE M G, BERARDI E.Evolution of sourdough microbiota in spontaneous sourdoughs started with different plant materials[J].International Journal of Food Microbiology, 2016, 232(6):35-42.
[2] 刘同杰.传统酸面团中微生物多样性及其风味物质代谢研究[D].杭州:浙江大学, 2018.
LIU T J.Biodiversity of Chinese traditional sourdough microbiota and its flavor metabolism[D].Hangzhou:Zhejiang University, 2018.
[3] 毛晋春.传统发酵酸面团微生物群落动态变化及其主要发酵菌株分离筛选[D].沈阳:沈阳农业大学, 2020.
MAO J C.Research on the dynamic changes of microbial community and the separation and screening of main fermenting strains in traditional fermented sourdough[D].Shenyang:Shenyang Agricultural University,2020.
[4] NIONELLIL, RIZZELLO C G.Sourdough-based biotechnologies for the production of gluten-free foods[J].Foods (Basel, Switzerland), 2016, 5(3):65.
[5] 杨紫璇,张宾乐, 蒋慧, 等.茅台酒曲植物乳杆菌大豆酸面团发酵面包的营养与抗氧化特性[J].食品与发酵工业, 2018, 44(6):37-42.
YANG Z X, ZHANG B L, JIANG H, et al.The nutrition and antioxidant properties of soybean sourdough and bread fermented by LAB screened from Maotai Qu starter[J].Food and Fermentation Industries, 2018, 44(6):37-42.
[6] DEBONNEE, VAN SCHOORS F, MAENE P, et al.Comparison of the antifungal effect of undissociated lactic and acetic acid in sourdough bread and in chemically acidified wheat bread[J].International Journal of Food Microbiology, 2020, 321:108551.
[7]  SPIRLIH, ÖZMEN D, Y
SPIRLIH, ÖZMEN D, Y LMAZ M T, et al.Impact of glucan type exopolysaccharide (EPS) production on technological characteristics of sourdough bread[J].Food Control, 2020, 107:106812.
LMAZ M T, et al.Impact of glucan type exopolysaccharide (EPS) production on technological characteristics of sourdough bread[J].Food Control, 2020, 107:106812.
[8] 尹艳丽,王金水, 蔺丹华, 等.酸面团和谷物发酵对面制食品营养品质的影响[J].食品与发酵工业, 2015, 41(10):230-234.
YIN Y L, WANG J S, LIN D H, et al.Influence of sourdough and cereal fermentation on nutritional quality of wheat flour foods[J].Food and Fermentation Industries, 2015, 41(10):230-234.
[9] DEVUYST L, VAN KERREBROECK S, HARTH H, et al.Microbial ecology of sourdough fermentations:Diverse or uniform?[J].Food Microbiology, 2014, 37:11-29.
[10] MINERVINIF, DE ANGELIS M, DI CAGNO R, et al.Ecological parameters influencing microbial diversity and stability of traditional sourdough[J].International Journal of Food Microbiology, 2014, 171:136-146.
[11] ROSSIS, PARROTTA L, DEL DUCA S, et al.Effect of Yarrowia lipolyticaRO25 cricket-based hydrolysates on sourdough quality parameters[J].LWT, 2021, 148:111760.
[12] OSHIROM, ZENDO T, NAKAYAMA J.Diversity and dynamics of sourdough lactic acid bacteriota created by a slow food fermentation system[J].Journal of Bioscience and Bioengineering, 2021, 131(4):333-340.
[13] G NZLEM G, ZHENG J S.Lifestyles of sourdough lactobacilli-Do they matter for microbial ecology and bread quality?[J].International Journal of Food Microbiology, 2019, 302:15-23.
NZLEM G, ZHENG J S.Lifestyles of sourdough lactobacilli-Do they matter for microbial ecology and bread quality?[J].International Journal of Food Microbiology, 2019, 302:15-23.
[14] DECOCKP, CAPPELLE S.Bread technology and sourdough technology[J].Trends in Food Science &Technology, 2005, 16(1):113-120.
[15] REALEA, DI RENZO T, PREZIUSO M, et al.Stabilization of sourdough starter by spray drying technique:New breadmaking perspective[J].LWT, 2019, 99:468-475.
[16] CAGLARN,ERMIS E, DURAK M Z.Spray-dried and freeze-dried sourdough powders:Properties and evaluation of their use in breadmaking[J].Journal of Food Engineering, 2021, 292:110355.
[17] GALLIV, VENTURI M, PINI N, et al.Liquid and firm sourdough fermentation:Microbial robustness and interactions during consecutive backsloppings[J].LWT, 2019, 105:9-15.
[18] XINGX L, MA J Y, FU Z J, et al.Diversity of bacterial communities in traditional sourdough derived from three terrain conditions (mountain, plain and basin) in Henan Province, China[J].Food Research International (Ottawa,Ont.), 2020, 133:109139.
[19] VRANCKENG, DE VUYST L, VAN DER MEULEN R, et al.Yeast species composition differs between artisan bakery and spontaneous laboratory sourdoughs[J].FEMS Yeast Research, 2010, 10(4):471-481.
[20] WANGY H, YANG Y Y, LI H Q, et al.Characterization of aroma-active compounds in steamed breads fermented with Chinese traditional sourdough[J].LWT, 2021, 152:112347.
[21] WUS M, PENG Y L, XI J Z, et al.Effect of sourdough fermented with corn oil and lactic acid bacteria on bread flavor[J].LWT, 2022, 155:112935.
[22] TANGT P, LI Q, HUANG Z W, et al.Evaluation of Shandong pancake with sourdough fermentation on the alleviation of type 2 diabetes symptoms in mice[J].Journal of Functional Foods, 2022, 90:104952.
[23] OZGOLETM, YAMAN M, ZEKI DURAK M, et al.The effect of five different sourdough on the formation of glyoxal and methylglyoxal in bread and influence ofin vitro digestion[J].Food Chemistry, 2022, 371:131141.
[24] R
 Y
Y OR, RUDY S, KRZYKOWSKI A, et al.Gluten-free bread prepared with fresh and freeze-dried rice sourdough-texture and sensory evaluation[J].Journal of Texture Studies, 2016, 47(5):443-453.
OR, RUDY S, KRZYKOWSKI A, et al.Gluten-free bread prepared with fresh and freeze-dried rice sourdough-texture and sensory evaluation[J].Journal of Texture Studies, 2016, 47(5):443-453.
[25] DEVUYST L, VANCANNEYT M.Biodiversity and identification of sourdough lactic acid bacteria[J].Food Microbiology, 2007, 24(2):120-127.
[26] 邢小龙.河南地区老酵面团菌群结构及优势菌种复配研究[D].郑州:河南农业大学, 2020.
XING X L.Microbial communities in Chinese traditional fermented dough from Henan Province and compound properties of predominant microorganism[D].Zhengzhou:Henan Agricultural University, 2020.
[27] 张国华,王伟, 涂建, 等.基于宏转录组学技术解析传统酸面团中微生物代谢机理[J].中国粮油学报, 2019, 34(11):10-16.
ZHANG G H, WANG W, TU J, et al.Analysis of microbial metabolism mechanism in traditional sourdough fermentation based on macro-transcriptomics technology[J].Journal of the Chinese Cereals and Oils Association, 2019, 34(11):10-16.
[28] DEVUYST L, VRANCKEN G, RAVYTS F, et al.Biodiversity, ecological determinants, and metabolic exploitation of sourdough microbiota[J].Food Microbiology, 2009, 26(7):666-675.
[29] ROCHAJ M, MALCATA F X.Microbiological profile of maize and rye flours, and sourdough used for the manufacture of traditional Portuguese bread[J].Food Microbiology, 2012, 31(1):72-88.
[30] HAMMESW P, BRANDT M J, FRANCIS K L, et al.Microbial ecologyof cereal fermentations[J].Trends in Food Science &Technology, 2005, 16(1-3):4-11.
[31] DINARDOF R, MINERVINI F, DE ANGELIS M, et al.Dynamics of Enterobacteriaceae and lactobacilli in model sourdoughs are driven by pH and concentrations of sucrose and ferulic acid[J].LWT, 2019, 114:108394.
[32] STEFANELLOR F, MACHADO A A R, PASQUALIN CAVALHEIRO C, et al.Trehalose as a cryoprotectant in freeze-dried wheat sourdough production[J].LWT, 2018, 89:510-517.
[33] DENTICEMAIDANA S D, ARISTIMU O FICOSECO C A, BASSI D, et al.Biodiversity and technological-functional potential of lactic acid bacteria isolated from spontaneously fermented chia sourdough[J].International Journal of Food Microbiology, 2020, 316:108425.
O FICOSECO C A, BASSI D, et al.Biodiversity and technological-functional potential of lactic acid bacteria isolated from spontaneously fermented chia sourdough[J].International Journal of Food Microbiology, 2020, 316:108425.
[34] OSHIROM, MOMODA R, TANAKA M, et al.Dense tracking of the dynamics of the microbial community and chemicals constituents in spontaneous wheat sourdough during two months of backslopping[J].Journal of Bioscience and Bioengineering, 2019, 128(2):170-176.
[35] WECKXS, VAN DER MEULEN R, ALLEMEERSCH J, et al.Community dynamics of bacteria in sourdough fermentations as revealed by their metatranscriptome[J].Applied and Environmental Microbiology, 2010, 76(16):5 402-5 408.
[36] VANDER MEULEN R, SCHEIRLINCK I, VAN SCHOOR A, et al.Population dynamics and metabolite target analysis of lactic acid bacteria during laboratory fermentations of wheat and spelt sourdoughs[J].Applied and Environmental Microbiology, 2007, 73(15):4 741-4 750.
[37] DEVUYST L, NEYSENS P.The sourdough microflora:Biodiversity and metabolic interactions[J].Trends in Food Science &Technology, 2005, 16(1-3):43-56.
[38] G NZLEM G, VERMEULEN N, VOGEL R F.Carbohydrate, peptide and lipid metabolism of lactic acid bacteria in sourdough[J].Food Microbiology, 2007, 24(2):128-138.
NZLEM G, VERMEULEN N, VOGEL R F.Carbohydrate, peptide and lipid metabolism of lactic acid bacteria in sourdough[J].Food Microbiology, 2007, 24(2):128-138.
[39] DEANGELIS M, BINI L C, PALLINI V, et al.The acid-stress response in Lactobacillus sanfranciscensisCB1[J].Microbiology (Reading, England), 2001, 147(Pt 7):1 863-1 873.
[40] VOGELR F, PAVLOVIC M, EHRMANN M A, et al.Genomic analysisreveals Lactobacillus sanfranciscensisas stable element in traditional sourdoughs[J].Microbial Cell Factories, 2011, 10(1):S6.
[41] VANKERREBROECK S, MAES D, DE VUYST L.Sourdoughs as a function of their species diversity and process conditions, a meta-analysis[J].Trends in Food Science &Technology, 2017, 68:152-159.
[42] GOBBETTIM, MINERVINI F, PONTONIO E, et al.Drivers for the establishment and composition of the sourdough lactic acid bacteria biota[J].International Journal of Food Microbiology, 2016, 239:3-18.
[43] MENEZESL A A, SARDARO M L S, DUARTE R T D, et al.Sourdough bacterial dynamics revealed by metagenomic analysis in Brazil[J].Food Microbiology, 2020, 85:103302.
[44] ÇAK RE, AR
RE, AR C
C M, DURAK M Z.Biodiversity and techno-functional properties of lactic acid bacteria in fermented hull-less barley sourdough[J].Journal of Bioscience and Bioengineering, 2020, 130(5):450-456.
M, DURAK M Z.Biodiversity and techno-functional properties of lactic acid bacteria in fermented hull-less barley sourdough[J].Journal of Bioscience and Bioengineering, 2020, 130(5):450-456.
[45] COMASIOA, VERCE M, VAN KERREBROECK S, et al.Diverse microbial composition of sourdoughs from different origins[J].Frontiers in Microbiology, 2020, 11:1212.
[46] FEKRIA, TORBATI M, YARI KHOSROWSHAHI A, et al.Functional effects of phytate-degrading, probiotic lactic acid bacteria and yeast strains isolated from Iranian traditional sourdough on the technological and nutritional properties of whole wheat bread[J].Food Chemistry, 2020, 306:125620.
[47] MORONIA V, ARENDT E K, DAL BELLO F.Biodiversity of lactic acid bacteria and yeasts in spontaneously-fermented buckwheat and teff sourdoughs[J].Food Microbiology, 2011, 28(3):497-502.
[48] MICHELE, MONFORT C, DEFFRASNES M, et al.Characterization of relative abundance of lactic acid bacteria species in French organic sourdough by cultural, qPCR and MiSeq high-throughput sequencing methods[J].International Journal of Food Microbiology, 2016, 239(11):35-43.
[49] BARTKIENEE, LELE V, RUZAUSKAS M, et al.Lactic acid bacteria isolation from spontaneous sourdough and their characterization including antimicrobial and antifungal properties evaluation[J].Microorganisms, 2019, 8(1):64.
[50] FRABERGERV, UNGER C, KUMMER C, et al.Insights into microbial diversity of traditional Austrian sourdough[J].LWT, 2020, 127:109358.
[51] RAIMONDIS, AMARETTI A, ROSSI M, et al.Evolution of microbial community and chemical properties of a sourdough during the production of Colomba, an Italian sweet leavened baked product[J].LWT, 2017, 86:31-39.
[52] CODAR, KIANJAM M, PONTONIO E, et al.Sourdough-type propagation of faba bean flour:Dynamics of microbial consortia and biochemical implications[J].International Journal of Food Microbiology, 2017, 248:10-21.
[53] YANGQ W, RUTHERFURD-MARKWICK K, MUTUKUMIRA A N.Identification of dominant lactic acid bacteria and yeast in rice sourdough produced in New Zealand[J].Current Research in Food Science, 2021, 4:729-736.
[54] HARTHH, VAN KERREBROECK S, DE VUYST L.Community dynamics and metabolite target analysis of spontaneous, backslopped barley sourdough fermentations under laboratory and bakery conditions[J].International Journal of Food Microbiology, 2016, 228:22-32.
[55] FUJIMOTOA, ITO K, NARUSHIMA N, et al.Identification of lactic acid bacteria and yeasts, and characterization of food components of sourdoughs used in Japanese bakeries[J].Journal of Bioscience and Bioengineering, 2019, 127(5):575-581.
[56] LIUT J, LI Y, CHEN J C, et al.Prevalence and diversity of lactic acid bacteria in Chinese traditional sourdough revealed by culture dependent and pyrosequencing approaches[J].LWT-Food Science and Technology, 2016, 68:91-97.
[57] YANB W, SADIQ F A, CAI Y J, et al.Microbial diversity in traditional type I sourdough and Jiaozi and its influence on volatiles in Chinese steamed bread[J].LWT, 2019, 101:764-773.
[58] YANGH Y, LIU T J, ZHANG G H, et al.Intraspecific diversity and fermentative properties of Saccharomyces cerevisiaefrom Chinese traditional sourdough[J].LWT, 2020, 124:109195.
[59] BARTLEL, SUMBY K, SUNDSTROM J, et al.The microbial challenge of winemaking:Yeast-bacteria compatibility[J].FEMS Yeast Research, 2019, 19(4):foz040.
[60] 姚尚杰,金垚, 周荣清, 等.传统发酵食品中微生物间相互作用及应用[J].生物产业技术, 2019(4):48-54.
YAO S J, JIN Y, ZHOU R Q, et al.Interaction and its application of microorganisms in traditional fermented food[J].Biotechnology &Business, 2019(4):48-54.
[61] GOBBETTIM.The sourdough microflora:Interactions of lactic acid bacteria and yeasts[J].Trends in Food Science &Technology, 1998, 9(7):267-274.
[62] LIMP L, TOH M, LIU S Q.Saccharomyces cerevisiaeEC—1118 enhances the survivability of probiotic Lactobacillus rhamnosus HN001 in an acidic environment[J].Applied Microbiology and Biotechnology, 2015, 99(16):6 803-6 811.
[63] SIEUWERTSS, BRON P A, SMID E J.Mutually stimulating interactions between lactic acid bacteria and Saccharomyces cerevisiaein sourdough fermentation[J].LWT, 2018, 90:201-206.
[64] 乌云,高增丽, 曹文慧, 等.传统列巴面团中潜在共生菌的筛选[J].中国食品添加剂, 2020, 31(10):85-94.
WU Y, GAO Z L, CAO W H, et al.Screening of potential symbiotic bacteria in traditional leba dough[J].China Food Additives, 2020, 31(10):85-94.
[65] WINTERSM, PANAYOTIDES D, BAYRAK M, et al.Defined co-cultures of yeast and bacteria modify the aroma, crumb and sensory properties of bread[J].Journal of Applied Microbiology, 2019, 127(3):778-793.
[66] BOUDAOUDS, AOUF C, DEVILLERS H, et al.Sourdough yeast-bacteria interactions can change ferulic acid metabolism during fermentation[J].Food Microbiology, 2021, 98:103790.
[67] NARVHUSJ A, GADAGA T H.The role of interaction between yeasts and lactic acid bacteria in African fermented milks:A review[J].International Journal of Food Microbiology, 2003, 86(1-2):51-60.
[68] SUIHKOM L, M KINEN V.Tolerance of acetate, propionate and sorbate by Saccharomyces cerevisiaeand Torulopsis holmii[J].Food Microbiology, 1984, 1(2):105-110.
KINEN V.Tolerance of acetate, propionate and sorbate by Saccharomyces cerevisiaeand Torulopsis holmii[J].Food Microbiology, 1984, 1(2):105-110.
[69] MINERVINIF, LATTANZI A, DINARDO F R, et al.Wheat endophytic lactobacilli drive the microbial and biochemical features of sourdoughs[J].Food Microbiology, 2018, 70:162-171.
[70] BRANDTM J, HAMMES W P, G NZLE M G.Effects of process parameters on growth and metabolism of Lactobacillus sanfranciscensis andCandida humilisduring rye sourdough fermentation[J].European Food Research and Technology, 2004, 218(4):333-338.
NZLE M G.Effects of process parameters on growth and metabolism of Lactobacillus sanfranciscensis andCandida humilisduring rye sourdough fermentation[J].European Food Research and Technology, 2004, 218(4):333-338.
[71] YANGH Y, SADIQ F A, LIU T J, et al.Use of physiological and transcriptome analysis to infer the interactions between Saccharomyces cerevisiaeand Lactobacillus sanfranciscensisisolated from Chinese traditional sourdoughs[J].LWT, 2020, 126:109268.
[72] VOGELMANNS A, SEITTER M, SINGER U, et al.Adaptability of lactic acid bacteria and yeasts to sourdoughs prepared from cereals, pseudocereals and cassava and use of competitive strains as starters[J].International Journal of Food Microbiology, 2009, 130(3):205-212.
[73] LANDISE A, OLIVERIO A M, MCKENNEY E A, et al.The diversity and function of sourdough starter microbiomes[J].eLife, 2021, 10:e61644.
[74] LIH F, FU J K, HU S, et al.Comparison of the effects of acetic acid bacteria and lactic acid bacteria on the microbial diversity of and the functional pathways in dough as revealed by high-throughput metagenomics sequencing[J].International Journal of Food Microbiology, 2021, 346:109168.
[75] SAICHANAN, MATSUSHITA K, ADACHI O, et al.Acetic acid bacteria:A group of bacteria with versatile biotechnological applications[J].Biotechnology Advances, 2015, 33(6):1 260-1 271.
[76] GOMESR J, BORGES M F, ROSA M F, et al.Acetic acid bacteria in the food industry:Systematics, characteristics and applications[J].Food Technology and Biotechnology, 2018, 56(2):139-151.