玉米赤霉烯酮(zearalenone,ZEN),又称F-2毒素,主要由禾谷镰刀菌(Fusarium graminearum)等镰刀菌属真菌产生的有毒代谢产物[1],是一种经聚酮途径生物合成的非甾体雌激素类真菌毒素,食用被ZEN污染的食品将危害人和动物的身体健康(图1)。本文将简要介绍ZEN的危害及污染现状,系统总结ZEN的生物转化机理、降解酶的挖掘及分子改造研究进展。
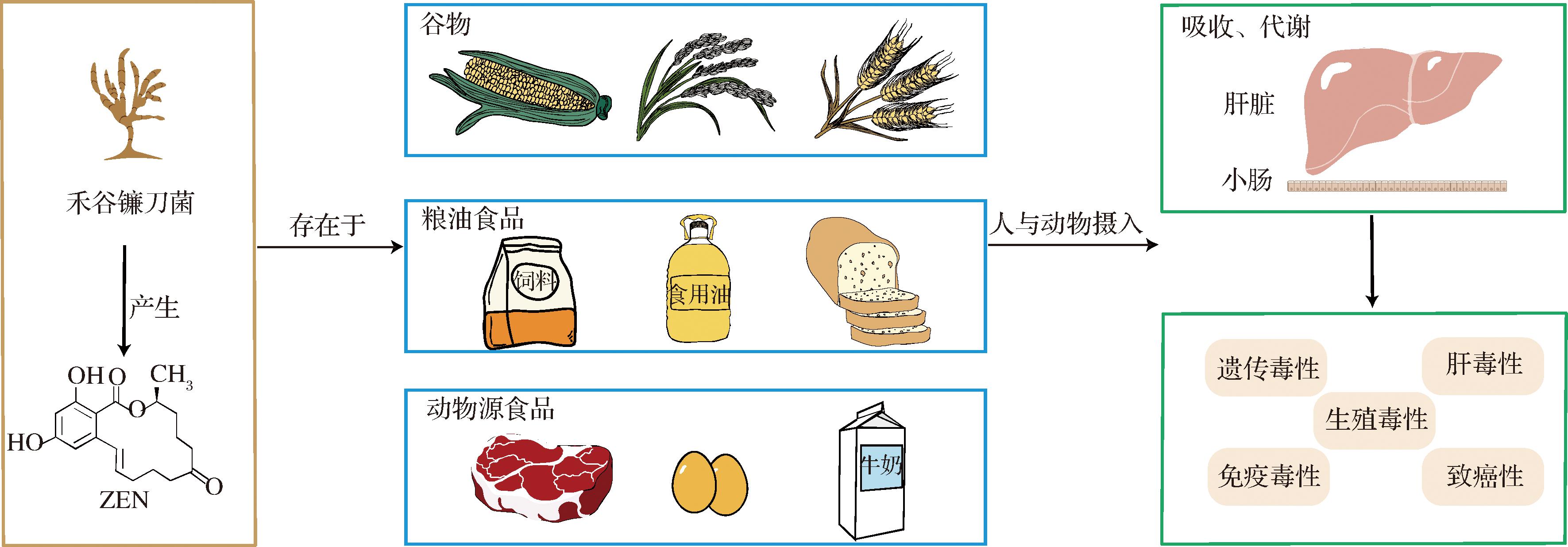
图1 食品中ZEN污染及对人畜健康的危害
Fig.1 ZEN pollution in food and its harm to human and animal health
1 ZEN及其衍生物的危害
ZEN分子式为C18H22O5,具有二羟基苯甲酸内酯结构,不溶于水,易溶于有机溶剂,结构稳定,具有较强耐热性。ZEN结构与雌激素相似,可通过与雌激素受体(estrogen receptor,ER)竞争性结合而影响人和动物的内分泌系统,摄入被ZEN及其衍生物污染的食物,经肝脏和肠道吸收、代谢后,将造成生殖、免疫、遗传毒性与致癌性[2]。此外,ZEN还可通过诱导氧化损伤和应激蛋白的过度表达产生细胞毒性[3],ZEN诱导产生的氧化应激和脂质过氧化还可导致肝毒性[4]。ZEN及其衍生物(图2)可由产毒真菌直接产生,ZEN经植物、动物、微生物转化后可生成玉米赤霉酮(zearalanone,ZAN)、α-玉米赤霉烯醇(α-zearalenol,α-ZOL)和β-玉米赤霉烯醇(β-zearalenol,β-ZOL),α-ZOL和β-ZOL还可进一步还原为α-玉米赤霉醇(α-zearalanol,α-ZAL)和β-玉米赤霉醇(β-zearalanol,β-ZAL)[5]。另外,ZEN也能转化为毒性更低的衍生物玉米赤霉烯酮-14-硫酸盐(zearalenone-14-Sulfate,Z14S)和玉米赤霉烯酮-14-葡萄糖苷(zearalenone-14-glucoside,Z14G)。其中,α-ZOL的毒性最高,其雌激素活性是ZEN的16倍,α-ZAL的雌激素活性是ZEN的7倍[6]。我国在GB 2761—2017 《食品安全国家标准 食品中真菌毒素限量》规定谷物及其制品中ZEN的限量为60 μg/kg;欧盟委员会在(EU)2023/915对ZEN限量进行了详细分类,规定谷物及其制品的限量为50~100 μg/kg;联合国粮食及农业组织和世界卫生组织食品添加剂联合专家委员会将ZEN的每日最大耐受摄入量暂定为0.5 μg/kg bw[7]。
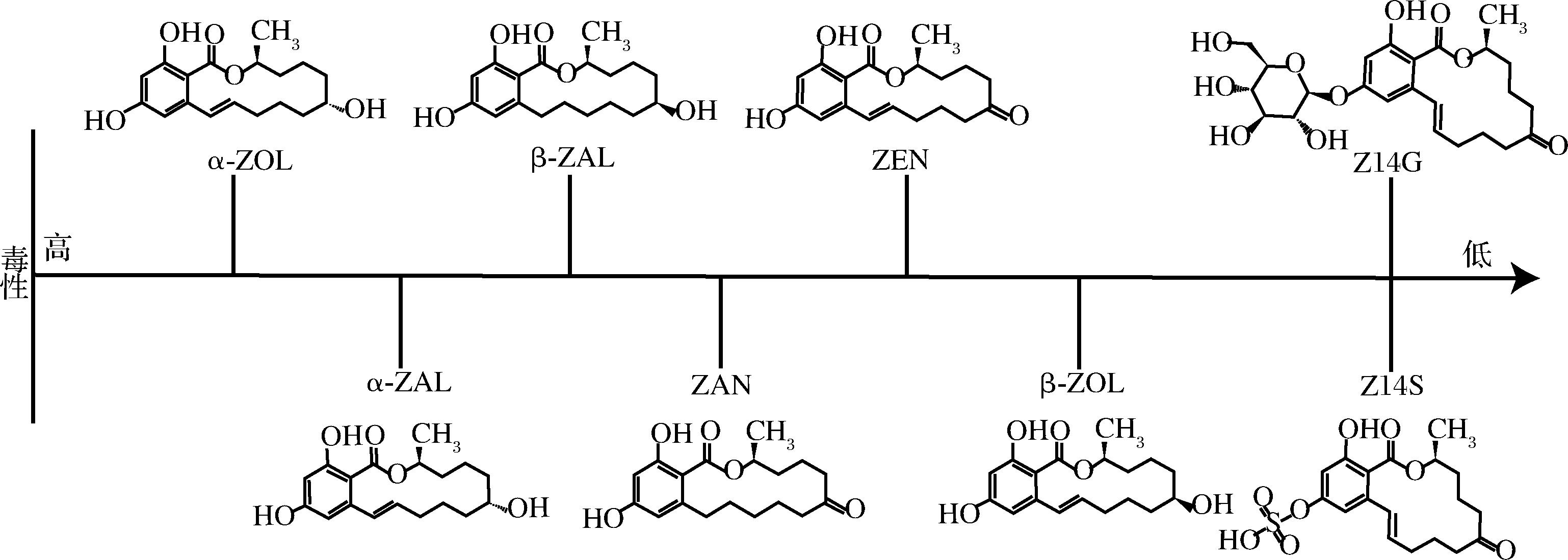
图2 ZEN及其衍生物的结构
Fig.2 Structure of ZEN and its derivatives
2 玉米赤霉烯酮的污染现状
ZEN不仅可广泛污染玉米、小麦、大米、大麦、高粱、大豆、燕麦等谷物,还可以ZEN及其衍生物的形式存在于肉、蛋、奶等动物源性食品,以及食用油、面包、啤酒、玉米蛋白粉、酒糟蛋白等谷物来源的粮油食品和饲料原料中[8]。近年来,全球ZEN的污染形势严峻,其中以温带气候地区,特别是欧洲、亚洲和北美部分地区尤为严重。2021年对采自世界各地的饲料原料、玉米、小麦、副产品等样品分析结果发现,ZEN的检出率达到79%,而2020年该值仅为47%[9]。我国也是ZEN污染较为严重的国家之一,其中以华东、华南、华北地区的污染情况较为突出。2021年对我国部分地区饲料原料的调查结果显示,ZEN检出率达到82.2%,其中,玉米中ZEN的检出率达到79.1%[10]。2022年对江西省饲料及饲料原料调查发现,ZEN的检出率为74.4%,其中,饲料原料中酒糟类检出率最高,达到87.5%[11]。此外,ZEN可在谷物加工过程中进一步富集,比如,玉米毛油中ZEN的污染情况就非常严峻,裴娅晓等[12]从玉米油加工厂取得的玉米毛油中ZEN含量高达8 153.33 μg/kg。ZHAO等[13]从中国不同玉米油厂获得的脱胶玉米油中ZEN的含量最高可达7 066 μg/kg。
3 玉米赤霉烯酮的生物脱毒
近年来,随着对ZEN结构及毒性研究的不断深入,针对食品及饲料中ZEN的污染情况,人们提出了许多脱毒策略,主要分为物理、化学和生物脱毒方法。其中,物理方法包括研磨[14]、筛分与脱壳[15]、热处理[16]、辐射[17]、高压处理[18]、吸附[19]等。化学方法是利用臭氧、焦亚硫酸钠、甲胺、氢氧化钙、亚硫酸钠、环糊精、植物提取物等与ZEN发生碱化、水解、氧化和还原反应[20-23]。这两类方法靶向性差,在去除ZEN的同时也存在脱毒效果不稳定、营养物质损失、化学物质二次污染等诸多问题。
生物脱毒方法以生物吸附和生物降解为主要途径,生物吸附是利用微生物细胞壁上的特殊结构与ZEN在一定条件下以氢键、离子键和疏水相互作用等方式结合,从而实现ZEN的吸附去除[24]。其中,乳酸菌细胞壁的蛋白和肽聚糖的去质子化羧基[25]与酵母细胞壁的β-1,3-葡聚糖和β-1,6-葡聚糖组成的三维结构在吸附机制中起着重要作用[26-27]。但该法并未破坏毒素结构,使用中需控制条件,防止因解吸附而造成质量安全风险。生物降解是ZEN经功能性微生物产生的生物酶催化进而转化成无毒或低毒产物[28]。此过程一般通过ZEN二羟基苯环外部结构的硫基化、羟基化、糖基化、磷酸化和内酯环的破坏来实现,该策略因其绿色、安全、高效、不会造成营养成分损失等诸多优势受到国内外研究学者的广泛关注。目前,已知的ZEN降解酶种类有限,且部分降解机理尚不明晰。因此,本文综合ZEN的降解机理及降解酶的分类进行归纳总结,以期为新型ZEN降解酶的挖掘提供理论及应用参考。
3.1 水解酶
ZEN水解酶(zearalenone hydrolase,ZHD)是ZEN生物降解中研究最为广泛且深入的一类降解酶,目前已发现的水解酶及其性质如表1所示。EL-SHARKAWY等[29]研究发现粉红粘帚霉(Gliocladium roseum)NRRL 1859可裂解ZEN中的内酯键。2002年,首次从G.roseum IFO 7063中分离到可断开ZEN内酯键的酶[30],经过纯化、鉴定,克隆出了编码基因zhd101,并在大肠杆菌(Escherichia coli)[31]、水稻愈伤组织、酿酒酵母(Saccharomyces cerevisiae)[32]、罗伊氏乳杆菌(Lactobacillus reuteri)Pg4[33]等宿主中实现了异源表达。该酶降解机理为打开ZEN内酯环上的酯键形成中间产物,随后脱羧(图3),该产物为目前报道的毒性最低的ZEN降解产物。程波财等[34]成功从G.roseum中克隆出与ZHD101有9个碱基差异的DNA片段ZEN-jjm,经E.coli表达的重组酶细胞裂解上清液可在3 h内将液体中1 μg/mL的ZEN完全去除。刘海燕等[35]同样从中克隆出了与ZHD101有11个碱基差异的降解酶基因zlhy-6,在毕赤酵母中表达后也可降解液体中的ZEN。经异源优化表达,该酶可将玉米油中ZEN降低至13 μg/mL,降解率达到96.31%[36]。
表1 不同ZEN水解酶的来源、异源表达及性质
Table 1 Source, heterologous expression, and properties of different ZEN hydrolases
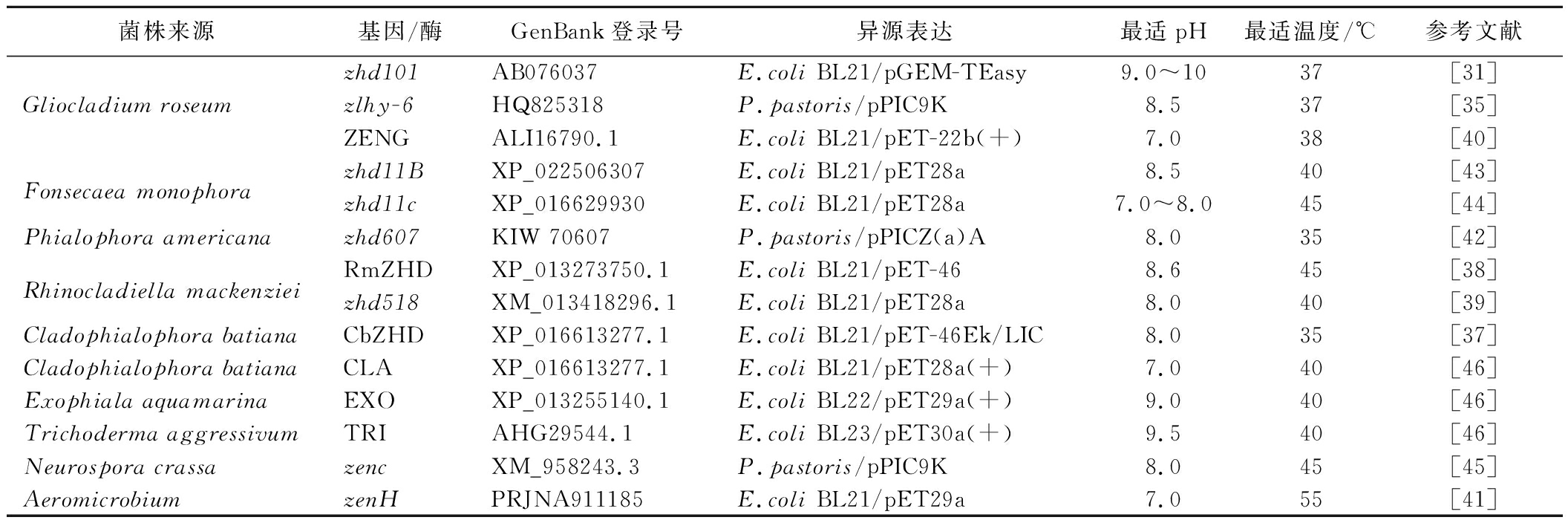
菌株来源基因/酶GenBank登录号异源表达最适pH最适温度/℃参考文献zhd101AB076037E.coli BL21/pGEM-TEasy9.0~1037[31]Gliocladium roseumzlhy-6HQ825318P.pastoris/pPIC9K8.537[35]ZENGALI16790.1E.coli BL21/pET-22b(+)7.038[40]Fonsecaea monophorazhd11BXP_022506307E.coli BL21/pET28a8.540[43]zhd11cXP_016629930E.coli BL21/pET28a7.0~8.045[44]Phialophora americanazhd607KIW 70607P.pastoris/pPICZ(a)A8.035[42]Rhinocladiella mackenzieiRmZHDXP_013273750.1E.coli BL21/pET-468.645[38]zhd518XM_013418296.1E.coli BL21/pET28a8.040[39]Cladophialophora batianaCbZHDXP_016613277.1E.coli BL21/pET-46Ek/LIC8.035[37]Cladophialophora batianaCLAXP_016613277.1E.coli BL21/pET28a(+)7.040[46]Exophiala aquamarinaEXOXP_013255140.1E.coli BL22/pET29a(+)9.040[46]Trichoderma aggressivumTRIAHG29544.1E.coli BL23/pET30a(+)9.540[46]Neurospora crassazencXM_958243.3P.pastoris/pPIC9K8.045[45]AeromicrobiumzenHPRJNA911185E.coli BL21/pET29a7.055[41]
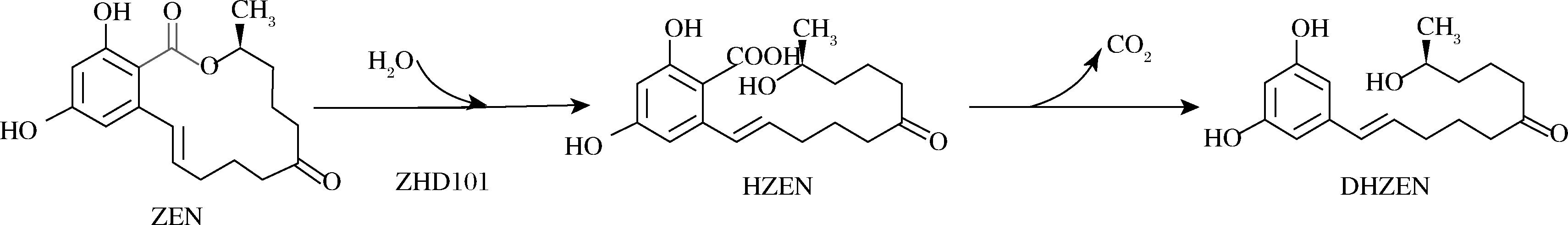
图3 水解酶ZHD101的降解机理
Fig.3 The degradation mechanism of hydrolase ZHD101
此外,研究人员还挖掘和研究了其他微生物来源的ZHD101同源蛋白,来自Cladophialophora batiana、Rhinocladiella mackenziei的水解酶CbZHD[37]、RmZHD[38]与Zhd518[39]对ZHD101有60%以上的同源性,其中Zhd518对ZEN的降解活性高达207.0 U/mg,并对其多个衍生物都有降解活性。ZHANG等[40]研究表明降解酶ZENG在中性条件下对ZEN及衍生物α-ZOL和α-ZAL也具有较高的降解活性,分别为315、187、117 U/mg。HU等[41]发现气微菌属(Aeromicrobium)HA中的降解酶ZenH在中性条件下的酶活力是ZENG的7倍,通过HR-ESI-MS测定了其降解产物P1(图4-a),推测该酶在将ZEN的酯键打开后,羧基进一步与酚羟基形成内酯。以RmZHD与Zhd518的氨基酸序列为基础,YU等[42]筛选出与RmZHD有74%同源性的ZHD607,ZEN的降解活性为82.333 U/mg。还有研究发现了与Zhd518有较高同源性的水解酶ZHD11B[43]和ZHD11C[44],前者在30~50 ℃内,保持至少60%的活性,后者在45 ℃孵育60 h后,仍保留近90%的活性。BI等[45]发现粗糙脉孢菌(Neurospora crassa)中的zenc基因经毕赤酵母(Pichia pastoris)高效表达后,ZENC(800 U)粗酶与含ZEN的酒糟蛋白、玉米副产物和玉米糠(25 g)共培养,可使ZEN含量分别降低70.9%、88.9%和94.7%。
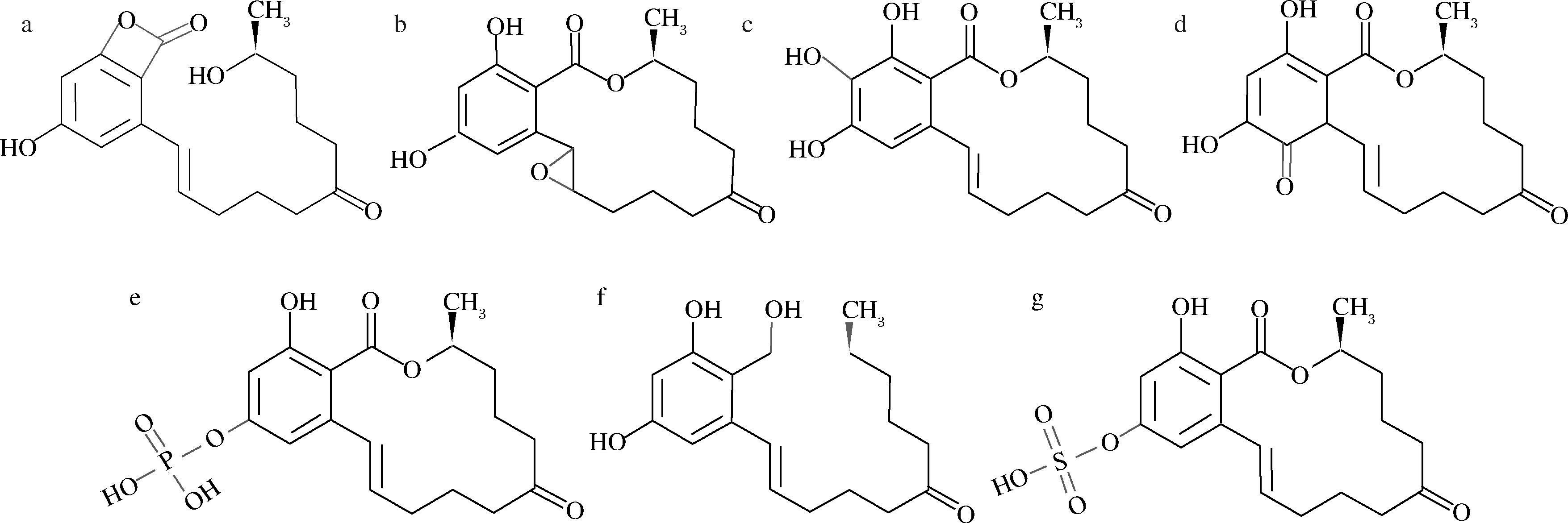
a-P1;b-ZEN-11,12-氧化物;c-15-OH-ZEN;d-13-OH-ZEN酮;e-ZEN-14-P;f-FSZ-P;g-Z14S
图4 其他降解产物结构
Fig.4 Structure of other degradation products
3.2 过氧化物酶
过氧化物酶是一种氧化还原酶,以H2O2作为电子受体实现对酚类底物的氧化,其对ZEN的降解作用已取得了一些进展(表2)。YU等[47]从不动杆菌(Acinetobacter)SM04中分离出的编码基因prx由E.coli表达后,重组过氧化物酶Prx在0.09%(质量分数)H2O2存在下处理ZEN含量为1 000 μg/kg的玉米粉,6 h后降解率可达90%,且饲料营养价值和适口性没有明显损失。GUO等[48]发现来自B.subtilis 168的染料脱色过氧化物酶BsDyP以H2O2为电子受体可直接氧化83%的ZEN,通过UPLC-MS/MS分析提出氧化产物为ZEN-11,12-氧化物(图4-b)。QIN等[49]将来自热羧链霉菌(Streptomyces thermocarboxydus)41291的基因StDyP在E.coli过表达,得到的重组酶有着锰过氧化物酶和漆酶的催化特性,1 mmol/L的Mn2+或1-羟基苯并三唑可显著促进ZEN降解为低毒产物15-OH-ZEN和13-OH-ZEN-醌(图4-c、图4-d)。研究表明,H2O2作为电子受体在上述氧化还原反应降解ZEN的过程中是必需的,但难以规模化应用,因此,人们提出了利用葡萄糖氧化酶(glucose oxidase,GOD)催化β-D-葡萄糖氧化生成D-葡萄糖-δ-内酯和H2O2,通过GOD和过氧化物酶级联反应来降解ZEN,但BsDyP在葡萄糖和GOD存在的条件下,对缓冲液中ZEN(10 μg/mL)与玉米浆中ZEN(9.2 μg/mL)的最大降解率分别为61%与33%[48]。因此,为保证降解效果,在应用过程中应充分考虑降解酶在复杂基质中的底物选择性。
表2 漆酶与过氧化物酶的来源及降解特性
Table 2 Sources and degradation properties of laccase and peroxidase
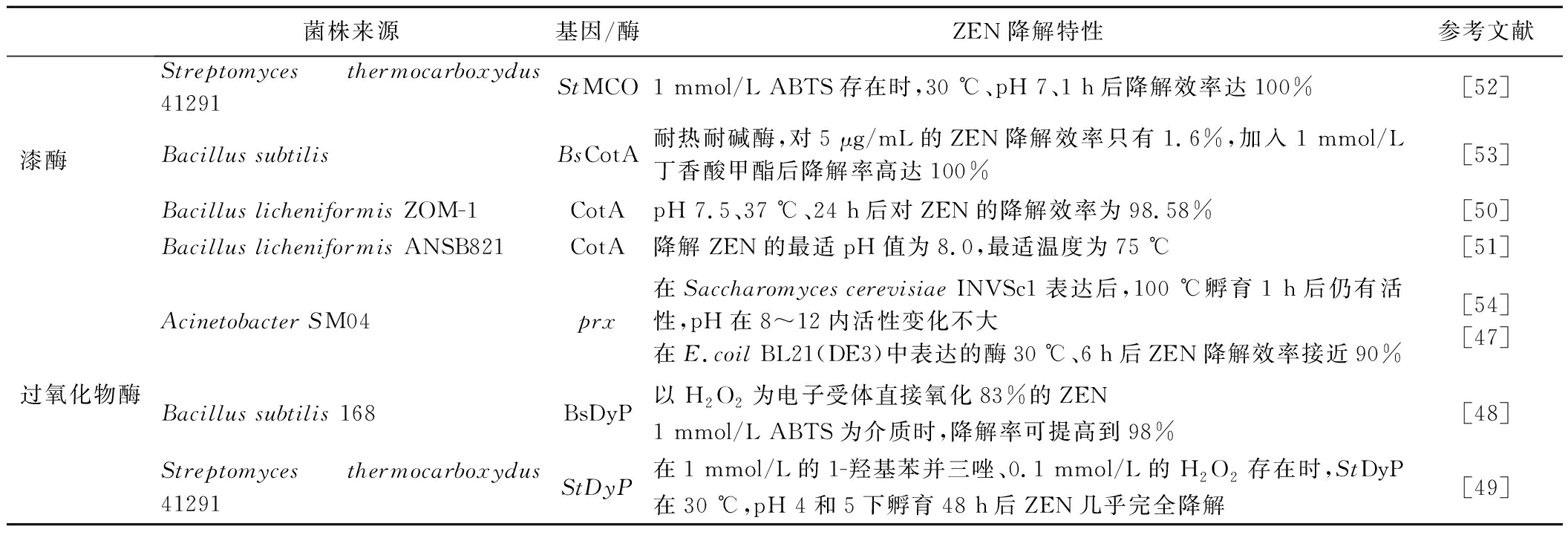
菌株来源基因/酶ZEN降解特性参考文献漆酶 Streptomyces thermocarboxydus 41291StMCO1 mmol/L ABTS存在时,30 ℃、pH 7、1 h后降解效率达100%[52]Bacillus subtilisBsCotA耐热耐碱酶,对5 μg/mL的ZEN降解效率只有1.6%,加入1 mmol/L丁香酸甲酯后降解率高达100%[53]Bacillus licheniformis ZOM-1CotApH 7.5、37 ℃、24 h后对ZEN的降解效率为98.58%[50]Bacillus licheniformis ANSB821CotA降解ZEN的最适pH值为8.0,最适温度为75 ℃[51]过氧化物酶Acinetobacter SM04prx在Saccharomyces cerevisiae INVSc1表达后,100 ℃孵育1 h后仍有活性,pH在8~12内活性变化不大在E.coil BL21(DE3)中表达的酶30 ℃、6 h后ZEN降解效率接近90%[54][47]Bacillus subtilis 168BsDyP以H2O2为电子受体直接氧化83%的ZEN1 mmol/L ABTS为介质时,降解率可提高到98%[48]Streptomyces thermocarboxydus 41291StDyP在1 mmol/L的1-羟基苯并三唑、0.1 mmol/L的H2O2存在时,StDyP在30 ℃,pH 4和5下孵育48 h后ZEN几乎完全降解[49]
3.3 漆酶
漆酶是一种含铜多酚氧化酶,以氧气为电子受体催化各种芳香族和非芳香族化合物的氧化,作为环保的生物催化剂广泛应用于食品、工业加工和生物能源开发等领域。研究表明,不同来源的漆酶对ZEN也表现出一定的降解特性(表2)。SUN等[50]从地衣芽孢杆菌(B.licheniformis)ZOM-1中克隆出的漆酶CotA,可将ZEN降解为低毒的15-OH-ZEN和13-OH-ZEN-醌。从B.licheniformis ANSB821中分离的漆酶CotA[51]与从S.thermocarboxydus 41291中分离的漆酶样多铜氧化酶StMCO[52],也有相似的降解途径。GUO等[51]还发现,用壳聚糖微球固定的漆酶CotA比游离酶具有更高的稳定性,从玉米粉溶液中回收、重复使用3次后,仍具有54%的降解率。漆酶降解ZEN可分为直接氧化和间接氧化,直接氧化可使ZEN的二羟基苯环部分羟基化,但在酸性条件下效率较低。间接氧化是通过添加的介质与漆酶形成自由基,自由基进一步与底物发生氧化反应,如B.licheniformis中CotA在酸性条件下,0.05 mmol/L的介质ABTS、乙酰丁香酮或丁香酸甲酯使其对ZEN的降解率可从15%提高到90%以上[51]。此外,从B.subtilis中分离的漆酶BsCotA添加1 mmol/L丁香酸甲酯后,可同时降解AFB1和ZEN[53]。
3.4 糖基转移酶
MICHLMAYR等[55]发现大麦UDP-葡萄糖基转移酶HvUGT14077可将ZEN降解为Z14G与玉米赤霉烯酮-16-O-葡萄糖苷(zearalenone-16-O-glucoside,Z16G),二者进一步结合为玉米赤霉烯酮-14,16-二葡糖苷(zearalenone-14,16-diglucoside,ZEN-14,16-diG),如图5所示,动力学分析表明,该反应释放ZEN的逆反应速率很低,不到1%。PAN等[56]进一步印证了这一结论,近平滑假丝酵母(Candida parapsilosis)ATCC 7330在添加葡萄糖后,细胞膜上的葡萄糖基转移酶可将ZEN转化成ZEN-14,16-diG,并且在体外模拟胃肠消化实验中没有被进一步分解成ZEN,此外,该酶还可转化β-ZOL、α-ZOL和ZAN等ZEN的衍生物。

图5 糖基转移酶的降解机理
Fig.5 The degradation mechanism of glycosyltransferases
3.5 磷酸转移酶
ZHU等[57]报道了B.pumilis S62-W在28 ℃,120 r/min条件下与25 μg的ZEN共培养24 h,可将其完全转化为玉米赤霉烯酮-14-磷酸盐(ZEN-14-phosphate,ZEN-14-P)(图4-e)。同年,YANG等[58]发现B.subtilis Y816共培养7 h可完全降解40 mg/L的ZEN,并通过转录组测序分析得到了其中发挥重要作用的磷酸转移酶(zearalenone phosphotransferase,ZPH),该酶在过量的ATP与Mg2+存在下,可将ZEN高效转化为ZEN-14-P,磷酸化产物的雌激素活性与初始物质相比明显降低。赵程程等[59]发现沙福芽孢杆菌(B.safensis)M7L4的活细胞可将ZEN降解为ZEN-14-P,结合降解机理及基因功能注释分析,从NCBI数据库中挖掘到ZEN磷酸转移酶编码基因BsaPUE,经E.coli克隆并表达的重组酶BsaPUE,可在10 mmol/L的ATP及20 mmol/L的Mg2+存在下、30 min内将10 μg/mL的ZEN全部降解。
3.6 其他ZEN降解酶
随着生物技术的蓬勃发展,越来越多具有ZEN降解功能的生物资源被筛选和挖掘出来。XU等[60]利用高通量测序技术对具有降解ZEN功能的解淀粉芽孢杆菌(B.amyloliquefaciens)H6进行转录组分析及系列试验发现,辅酶A硫代酯酶YBGC/FADM家族蛋白ZTE138可以降解ZEN。SUN等[61]从发酵大豆中筛选到1株黑曲霉(Aspergillus niger)FS10,该菌胞外分泌的生物酶FSZ可降解ZEN及其衍生物α-ZAL、β-ZOL和ZAN,经超高效液相色谱-四极杆飞行时间质谱联用技术分析表明,FSZ可能通过破坏ZEN的含氧内酯环从而实现ZEN的脱毒(图4-f)[62]。为了提高酶的热稳定性,研究人员还采用稻壳为载体开展了FSZ的固定化研究,结果表明,固定化酶在90 ℃下仍可去除70%的ZEN[63]。此外,还有多种微生物被发现具有ZEN的降解功能,比如在研究1株木霉(Trichoderma)对具有ZEN产毒能力的禾谷镰刀菌菌丝生长的抑制作用时发现,该菌可将ZEN转化为还原和硫酸化形式的Z14S(图4-g)和玉米赤霉烯醇-14-硫酸酯(zearalenol-14-sulfate,ZOL14S)[64]。吡啶红球菌(Rhodococcus pyridinivorans)K408可能通过破环ZEN的内酯环和二羟基苯环实现对ZEN的降解,降解率可达80%以上[65]。酵母毛孢霉菌(Trichosporon mycotoxinivorans)可将ZEN转化为无雌激素毒性的ZOM-1(图6)以实现对ZEN的降解[66]。从土壤、瘤胃等不同样品中还筛选到了假单胞菌Pseudomonas otitidis TH-N16[67]、P.alcaliphila TH-C1和P.plecoglossicida TH-L1[68],以及稻瘟菌属(Shinella)Z-25T[69],芽孢杆菌B.amyloliquefaciens ZDS-1[70]、B.subtilis ANSB01G[71-72]、B.subtilis 168和B.natto CICC 24640[73]等微生物都具有ZEN降解功能,但针对降解机理、产物结构和安全性,以及降解酶等研究的进展非常有限,亟需开展深入、系统的研究以推进脱毒产品和技术的开发及应用。
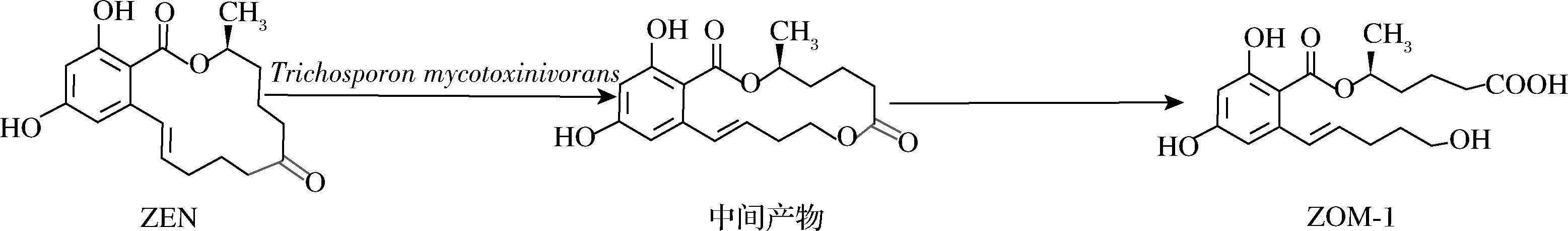
图6 酵母毛孢霉菌的降解机理
Fig.6 The degradation mechanism of Trichosporon mycotoxinivorans
4 ZEN降解酶的分子改造
粮油食品和饲料加工工艺中往往存在高温或极端pH等环境,目前报道的ZEN降解酶最适温度普遍为37~45 ℃,最适pH值为7~9,当温度升高至45 ℃或pH低于6时,酶活力迅速降低甚至丧失活性,且对毒性更高的α-ZOL的降解效果较差[74],在使用成本、热稳定性、底物选择性等方面无法满足工业化应用的需要。近年来,随着以蛋白质的结构模拟和分子动力学计算为基础的蛋白质分子设计的快速发展,ZEN降解酶在酶活力、热稳定性及底物特异性等方面取得了一些进展(表3)。
表3 ZEN降解酶的分子改造
Table 3 Molecular modification of ZEN degrading enzyme

菌株名称酶优化方法优化效果参考文献Gliocladium roseumZHD101ZLHY6ZENG定点突变(V153H)对α-ZOL的降解活性增加了3.7倍[75]定点突变(N156F、S194T和T259F)3个突变体酶活力相对于野生型分别为95.8%、131.6%和169.0%,双突变体N156F/S194T与三突变体N156F/S194T/T259F的Tm分别提高了6.7 ℃与6.1 ℃[76]定点突变(S220R和S220 W)2个突变体的Tm分别提高了5.6 ℃和4.0 ℃[77]定点突变(D157K和E171K)2个突变体的kcat/KM分别是野生型的1.34与2.06倍[78]引入二硫键(D143C/P181C)50 ℃处理2 min后的残余活性约为野生型的2倍[79]定点突变(H134 W)在45 ℃孵育20 min后,相对酶活力为野生型的10倍[80]引入二硫键(Q45C/A253C)在45 ℃孵育20 min后,相对酶活力为野生型的3.1倍定点突变(S162P/S220R)在55 ℃时比野生型酶活力提高了36.8倍[81]定点突变(H134F/S136F、H134I/S136I、H134L/S136L)3种突变体在48 ℃孵育超过7 min后保留了其40%的活性[40]Rhinoccladiella mack-enzieiZhd518定点突变(N156H)对α-ZOL的降解效率是野生型酶的3.3倍[39]Phialophora americanaZHD607定点突变(ZHDM1和I160Y)突变体ZHDM1和I160Y对ZEN的降解活性分别是ZHD607的2.9倍和3.4倍[42]Rhinocladiella mackenz-ieiRmZHD基因筛选有较高的耐热性,在pH 8.6和45 ℃条件下活性最高[38]定点突变(V153H和Y160A)V153H对α-ZOL的降解效率提高了3.17倍,Y160A对α-ZOL水解活性增加了70%Fonsecaea multimorpho-saZHD11C基因筛选在45 ℃孵育1 h后保留了近90%的活性,pH 6.5~9.0孵育12 h后仍保留超过12%的活性,能够水解α-ZAL、α-ZOL、β-ZAL、β-ZOL[44]Fonsecaea monophoraZhd11B基因筛选在30~50 ℃保持至少60%的相对活性,可以水解α-ZAL、α-ZOL、β-ZAL、β-ZOL、ZEN[43]帽子结构域交换(V131-L172替换为Zhd518的相应区域)对ZEN、α-ZAL和β-ZAL的活性分别提高了1.5、1.6、2.9倍[43]定点突变(T158H)对α-ZOL的相对活性增加了1.3倍[43]
4.1 提高降解酶活力
ZEN水解酶的晶体结构已逐渐被解析,ZHD101属于α/β-水解酶家族,由催化核心结构域与α-螺旋帽子结构域组成,底物结合在两者之间的深口袋中,与催化三联体Ser102-His242-Glu126相邻[82]。酶与底物的相互作用是影响酶活力的重要因素,改变结合口袋入口的构象、残基的刚性等可增加酶与底物的相互作用。通过晶体结构分析发现,降解酶RmZHD与ZHD101相比帽子结构域中的β6-α5环向内偏移,更接近底物结合口袋,且底物结合口袋的入口被Y160覆盖,呈现“闭合”构象,RmZHD中β6-α5环上N134介导的亲水接触和Y160介导的π堆积产生了额外底物相互作用可能是对ZEN降解效率较ZHD101提高了1.66倍的原因[38]。YU等[42]通过序列比对、结构分析与定点突变,筛选ZHD607中与ZEN降解活性相关的关键氨基酸,发现突变体ZHDM1和I160Y的比酶活分别较ZHD607提高了2.9倍和3.4倍,其中突变体ZHDM1的帽子结构域中,与底物口袋入口相对的环136~142显示出了较低的均方根波动值,帽子结构域的刚性提高,稳定了结合口袋中的底物;突变体I160Y与RmZHD的情况相似,催化口袋被Tyr160覆盖,“支撑”了ZEN结构,形成了一个更稳定的组合。此外,空间位阻也是影响酶活力的重要因素,ZHD101的第220位氨基酸处于结合口袋的上方,突变后侧链增大,可能导致结合口袋区域位置减小,增大底物进入活性区域的难度,使其ZEN降解活性降低[77]。
4.2 提高降解酶的耐酸性
改变pH活性曲线可使酶在偏离最佳pH情况下保持较高的活力,而改变酶的表面电荷可改变静电屏蔽和催化场效应进而影响pH活性曲线。LIN等[78]通过理性设计修改ZHD101中His242的静电环境(在蛋白质表面引入额外带正电的赖氨酸)来降低pKa值,从而使其pH活性曲线转移到酸性范围内,结果表明,突变体D157K与E171K在pH值为5.5时对ZEN保持较高的水解活性,kcat/KM分别是野生型的1.34与2.06倍。
4.3 提高降解酶热稳定性
蛋白质的结构与热稳定性密切相关,提升结构中柔性区域的刚性,可提高酶的热稳定性。而对于柔性区域的寻找,可借助分子动力学(molecular dynamics,MD)模拟和序列比对等方法。MD模拟可提供动态的分子信息,有助于蛋白结构中柔性位点的确定,避免盲目突变。FANG等[81]通过此方法获得了ZENG中2个均方根波动值最高的残基位点,在此柔性位点进行突变,筛选到了热稳定性提高的突变体S162P/S220R在55 ℃时比野生型酶活力提高了36.8倍,并发现突变体有额外氢键、盐桥的形成与脯氨酸的增加。用此方法还发现了ZLHY-6的热不稳定区段,对此区域的氨基酸进行虚拟饱和突变,突变体H134 W在45 ℃热处理20 min后,酶活力为野生型的10倍[80]。陈权等[76]基于MD模拟确定了突变位点并进行虚拟饱和突变,最后筛选得到的N156F、S194T和T259F三个突变体通过盐桥重排、NH-π的形成与蛋白表面空腔的填补来提高其热稳定性。除此之外,通过序列对比发现H134和S136对于大多数同源性较高的酶是保守的,但在热稳定性较高的RmZHD(I137和L139)中变化很大,ZHANG等[40]在ZENG的H134和S136位点设计了定点突变,成功获得了具有耐热性的突变体,在48 ℃孵育超过7 min后仍保留40%的酶活。官嫒林等[77]通过多蛋白的序列比对,在ZHD101的柔性区域进行虚拟饱和突变后,分析其序列保守性与构象自由能,最终得到突变体S220R和S220 W的Tm分别提高了5.6 ℃和4.0 ℃。
二硫键通过降低蛋白质解折叠速度和解折叠的主链熵值可稳定蛋白结构,因此,在酶的结构中引入二硫键也是提高热稳定性的有效手段。许中霞等[79]在ZHD101中通过双突变D143C/P181C引入二硫键,该突变体经50 ℃处理2 min后的残余活性约为野生型的2倍,但其在最适条件下的酶活力有所降低,为野生型的75%。杜昊澜等[80]使用Discovery Studio预测ZLHY-6中可突变形成二硫键的氨基酸后,用PyMol判断模拟后的结果,最后实验得到的突变体Q45C/A253C在45 ℃孵育20 min后的酶活力为野生型酶的3.1倍。
4.4 优化底物选择性
ZHD101降解α-ZOL的效率仅为ZEN的40%,分别将ZHD101与ZEN和α-ZOL复合物的晶体结构比对分析发现α-ZOL上C6’的羟基赋予α-ZOL内酯环更高的柔韧性,从而对ZHD101上亲水性的H242侧链产生排斥力,催化三联体之间形成的氢键网络被破坏,因此,与ZEN相比,对α-ZOL的降解效率较低,通过对酶-底物结合位点周围的几个氨基酸残基进行分子设计,得到的突变体V153H在保持ZEN降解活性的同时,对α-ZOL的催化活性增加了3.7倍[75]。利用分子动力学模拟和蛋白质结构分析发现153位的Val突变为His会引起ZHD101帽子结构域(Gly161~Thr190)的构象变化,从而增强酶-底物的氢键相互作用,使催化三联体中H242侧链向具有催化活性的位置移动,重构了催化三联体[83]。RmZHD、Zhd518和Zhd11B的突变体V153H、N156H和T158H对α-ZOL的降解效率分别提高了3.17、3.3、1.3倍[39,43]。值得注意的是,对于RmZHD来说,Y160的侧链虽然可增加与ZEN的相互作用,进而增加其降解活性,但对α-ZOL的C6’羟基有排斥作用,将此位点的氨基酸突变为Ala后,体积减小,降低了空间位阻,突变体Y160A对α-ZOL的水解活性提高了70%[38]。
2022年,从F.monophora中鉴定出了可水解ZEN、α-ZOL、β-ZOL、α-ZAL和β-ZAL的降解酶Zhd11B,但目前对其底物选择性的研究还不够深入,通过I-TASSER在线预测其晶体结构,发现帽子域结构中V131~L172位点与其他水解酶不同,将Zhd11B中这一区域替换为Zhd518的相应区域后,突变体Zhd11B-Zhd518对ZEN、α-ZAL和β-ZAL的活性分别提高了1.5、1.6、2.9倍[43]。
5 存在的问题和展望
ZEN的生物脱毒是近年来的研究热点,其中利用生物酶降解食品与饲料中的ZEN有很大应用前景,随着生物技术的不断发展,基于微生物多组学和蛋白质分子设计等策略的ZEN降解酶研究已经在新酶挖掘及酶的性能研究中取得了一些可喜的进展,为了更好地满足工业化生产和应用需求,未来还可从以下3个方面深入研究:a)目前已知的降解(菌)酶资源还较有限,因此,从自然界中挖掘和筛选仍然是获取ZEN降解生物资源的重要来源。通过高通量平台的降解微生物富集和选择性筛选,以及基于生物信息学比对和多组学分析的降解酶基因挖掘等方法均可有效获得具有降解功能的生物资源。b)采用蛋白质分子设计的酶性能改良可以极大地降低工作量、提高优化成功率,但目前基于计算机模拟和蛋白质分子设计挖掘新酶、提高酶催化特性的研究较有限,且该方法较依赖蛋白质的晶体结构,而针对未知分子结构的计算准确性还有待提高。基于此,结合酶与底物催化特征的构效关系进一步优化软件算法,提高酶结构、分子对接等预测的准确性,将成为降解酶研究的重要工具。c)工业化应用中需要降解酶具有性能稳定、使用成本低等优点,因此,可综合考虑酶催化活性和耐受性改良、底盘细胞设计、发酵和脱毒工艺优化等因素,以获得产量高、普适性好、生产和使用成本低的产品。
[1] BALL A, BUSZNY
A, BUSZNY KNÉ SZÉKV
KNÉ SZÉKV RI K, CZÉT
RI K, CZÉT NY P, et al. Estrogenic and non-estrogenic disruptor effect of zearalenone on male reproduction: A review[J]. International Journal of Molecular Sciences, 2023, 24(2):1578.
NY P, et al. Estrogenic and non-estrogenic disruptor effect of zearalenone on male reproduction: A review[J]. International Journal of Molecular Sciences, 2023, 24(2):1578.
[2] ZHENG W L, FENG N N, WANG Y, et al. Effects of zearalenone and its derivatives on the synthesis and secretion of mammalian sex steroid hormones: A review[J]. Food and Chemical Toxicology, 2019, 126:262-276.
[3] LIU X Y, XI H M, HAN S Q, et al. Zearalenone induces oxidative stress and autophagy in goat Sertoli cells[J]. Ecotoxicology and Environmental Safety, 2023, 252:114571.
[4] ZHANG C Q, LI C, LIU K C, et al. Characterization of zearalenone-induced hepatotoxicity and its mechanisms by transcriptomics in zebrafish model[J]. Chemosphere, 2022, 309:136637.
[5] MAHATO D K, DEVI S, PANDHI S, et al. Occurrence, impact on agriculture, human health, and management strategies of zearalenone in food and feed: A review[J]. Toxins, 2021, 13(2):92.
[6] STYPU A-TR
A-TR![]() BAS S, MINTA M, RADKO L, et al. Oestrogenic and (anti)androgenic activity of zearalenone and its metabolites in two in vitro yeast bioassays[J]. World Mycotoxin Journal, 2016, 9(2):247-256.
BAS S, MINTA M, RADKO L, et al. Oestrogenic and (anti)androgenic activity of zearalenone and its metabolites in two in vitro yeast bioassays[J]. World Mycotoxin Journal, 2016, 9(2):247-256.
[7] HAN X, HUANGFU B X, XU T X, et al. Research progress of safety of zearalenone: A review[J]. Toxins, 2022, 14(6):386.
[8] ROPEJKO K, ![]() M. Zearalenone and its metabolites-general overview, occurrence, and toxicity[J]. Toxins, 2021, 13(1):35.
M. Zearalenone and its metabolites-general overview, occurrence, and toxicity[J]. Toxins, 2021, 13(1):35.
[9] HAO W, LI A P, WANG J Y, et al. Mycotoxin contamination of feeds and raw materials in China in year 2021[J]. Frontiers in Veterinary Science, 2022, 9:929904.
[10] 刘建高, 厉学武, 彭哲, 等. 2021年我国饲料原料中常见霉菌毒素污染情况调查[J]. 中国家禽, 2023, 45(10):70-75.LIU J G, LI X W, PENG Z, et al. Investigation of common mycotoxins contamination in feed ingredients in China in 2021[J]. China Poultry, 2023, 45(10):70-75.
[11] 夏骏, 邢磊, 吴科盛, 等. 江西省饲料及饲料原料中真菌毒素污染水平调查与评估[J]. 饲料工业, 2024, 45(2):138-144.XIA J, XING L, WU K S, et al. Investigation and evaluation on mycotoxin contamination of raw materials and feed in Jiangxi Province[J]. Feed Industry, 2024, 45(2):138-144.
[12] 裴娅晓, 刘玉兰, 许利丽, 等. 碱炼脱除玉米油中玉米赤霉烯酮(ZEN)的研究[J]. 中国油脂, 2016, 41(5):56-60.PEI Y X, LIU Y L, XU L L, et al. Removal of zearalenone from maize oil by alkali refining[J]. China Oils and Fats, 2016, 41(5):56-60.
[13] ZHAO C W, XIE P K, JIN J, et al. Removal of zearalenone from degummed corn oil by hydrolase on a batch-refining unit[J]. Foods, 2022, 11(23):3795.
[14] LEE U S, JANG H S, TANAKA T, et al. Effect of milling on decontamination of Fusarium mycotoxins nivalenol, deoxynivalenol and zearalenone in Korean wheat[J]. Journal of Agricultural and Food Chemistry, 1987, 35(1):126-129.
[15] TRENHOLM H L, CHARMLEY L L, PRELUSKY D B, et al. Two physical methods for the decontamination of four cereals contaminated with deoxynivalenol and zearalenone[J]. Journal of Agricultural and Food Chemistry, 1991, 39(2):356-360.
[16] RYU D, HANNA M A, ESKRIDGE K M, et al. Heat stability of zearalenone in an aqueous buffered model system[J]. Journal of Agricultural and Food Chemistry, 2003, 51(6):1746-1748.
[17] LUO X H, QI L J, LIU Y T, et al. Effects of electron beam irradiation on zearalenone and ochratoxin A in naturally contaminated corn and corn quality parameters[J]. Toxins, 2017, 9(3):84.
[18] KALAGATUR N K, KAMASANI J R, MUDILI V, et al. Effect of high pressure processing on growth and mycotoxin production of Fusarium graminearum in maize[J]. Food Bioscience, 2018, 21:53-59.
[19] DU Q L, ZHANG W, XU N, et al. Efficient and simultaneous removal of aflatoxin B1, B2, G1, G2, and zearalenone from vegetable oil by use of a metal-organic framework absorbent[J]. Food Chemistry, 2023, 418:135881.
[20] YANG K, LI K, PAN L H, et al. Effect of ozone and electron beam irradiation on degradation of zearalenone and ochratoxin A[J]. Toxins, 2020, 12(2):138.
[21] SANTOS ALEXANDRE A P, VELA-PAREDES R S, SANTOS A S, et al. Ozone treatment to reduce deoxynivalenol (DON) and zearalenone (ZEN) contamination in wheat bran and its impact on nutritional quality[J]. Food Additives &Contaminants. Part A, Chemistry, Analysis, Control, Exposure &Risk Assessment, 2018, 35(6):1189-1199.
[22] PERCZAK A, ![]() K, et al. Degradation of zearalenone by essential oils under in vitro conditions[J]. Frontiers in Microbiology, 2016, 7:1224.
K, et al. Degradation of zearalenone by essential oils under in vitro conditions[J]. Frontiers in Microbiology, 2016, 7:1224.
[23] REMPE I, BREZINA U, KERSTEN S, et al. Effects of a Fusarium toxin-contaminated maize treated with sodium metabisulphite, methylamine and calcium hydroxide in diets for female piglets[J]. Archives of Animal Nutrition, 2013, 67(4):314-329.
[24] LUO Y, LIU X J, YUAN L, et al. Complicated interactions between bio-adsorbents and mycotoxins during mycotoxin adsorption: Current research and future prospects[J]. Trends in Food Science &Technology, 2020, 96:127-134.
[25] KR L A, POMASTOWSKI P,
L A, POMASTOWSKI P, ![]() K, et al. Microbiology neutralization of zearalenone using Lactococcus lactis and Bifidobacterium sp.[J]. Analytical and Bioanalytical Chemistry, 2018, 410(3):943-952.
K, et al. Microbiology neutralization of zearalenone using Lactococcus lactis and Bifidobacterium sp.[J]. Analytical and Bioanalytical Chemistry, 2018, 410(3):943-952.
[26] YIANNIKOURIS A, FRANÇOIS J, POUGHON L, et al. Alkali extraction of beta-d-glucans from Saccharomyces cerevisiae cell wall and study of their adsorptive properties toward zearalenone[J]. Journal of Agricultural and Food Chemistry, 2004, 52(11):3666-3673.
[27] FREIMUND S, SAUTER M, RYS P. Efficient adsorption of the mycotoxins zearalenone and T-2 toxin on a modified yeast glucan[J]. Journal of Environmental Science and Health, Part B, 2003, 38(3):243-255.
[28] 张静, 张琼琼, 计成, 等. 微生物及生物酶对脱氧雪腐镰刀菌烯醇生物转化研究进展[J]. 动物营养学报, 2020, 32(10):4807-4820.ZHANG J, ZHANG Q Q, JI C, et al. Research advance on biotransformation of deoxynivalenol by microbes and biological enzymes[J]. Chinese Journal of Animal Nutrition, 2020, 32(10):4807-4820.
[29] EL-SHARKAWY S, ABUL-HAJJ Y J. Microbial cleavage of zearalenone[J]. Xenobiotica, 1988, 18(4):365-371.
[30] KAKEYA H, TAKAHASHI-ANDO N, KIMURA M, et al. Biotransformation of the mycotoxin, zearalenone, to a non-estrogenic compound by a fungal strain of Clonostachys sp.[J]. Bioscience, Biotechnology, and Biochemistry, 2002, 66(12):2723-2726.
[31] TAKAHASHI-ANDO N, KIMURA M, KAKEYA H, et al. A novel lactonohydrolase responsible for the detoxification of zearalenone: Enzyme purification and gene cloning[J]. The Biochemical Journal, 2002, 365(Pt 1):1-6.
[32] TAKAHASHI-ANDO N, OHSATO S, SHIBATA T, et al. Metabolism of zearalenone by genetically modified organisms expressing the detoxification gene from Clonostachys rosea[J]. Applied and Environmental Microbiology, 2004, 70(6):3239-3245.
[33] YANG W C, HSU T C, CHENG K C, et al. Expression of the Clonostachys rosea lactonohydrolase gene by Lactobacillus reuteri to increase its zearalenone-removing ability[J]. Microbial Cell Factories, 2017, 16(1):69.
[34] 程波财, 史文婷, 罗洁, 等. 玉米赤霉烯酮降解酶基因(ZEN-jjm)的克隆、表达及活性分析[J]. 农业生物技术学报, 2010, 18(2):225-230.CHENG B C, SHI W T, LUO J, et al. Cloning of zearalenone-degraded enzyme gene (ZEN-jjm) and its expression and activity analysis[J]. Journal of Agricultural Biotechnology, 2010, 18(2):225-230.
[35] 刘海燕, 孙长坡, 伍松陵, 等. 玉米赤霉烯酮毒素降解酶基因的克隆及在毕赤酵母中的高效表达[J]. 中国粮油学报, 2011, 26(5):12-17.LIU H Y, SUN C P, WU S L, et al. Expression of zearalenone degrading enzyme gene from Gliocladium roseum in Pichia pastoris GS115[J]. Journal of the Chinese Cereals and Oils Association, 2011, 26(5):12-17.
[36] CHANG X J, LIU H J, SUN J, et al. zearalenone removal from corn oil by an enzymatic strategy[J]. Toxins, 2020, 12(2):117.
[37] HUI R J, HU X Y, LIU W T, et al. Characterization and crystal structure of a novel zearalenone hydrolase from Cladophialophora bantiana[J]. Acta Crystallographica Section F, 2017, 73(9):515-519.
[38] ZHENG Y Y, LIU W T, CHEN C C, et al. Crystal structure of a mycoestrogen-detoxifying lactonase from Rhinocladiella mackenziei: Molecular insight into ZHD substrate selectivity[J]. ACS Catalysis, 2018, 8(5):4294-4298.
[39] WANG M X, YIN L F, HU H Z, et al. Expression, functional analysis and mutation of a novel neutral zearalenone-degrading enzyme[J]. International Journal of Biological Macromolecules, 2018, 118:1284-1292.
[40] ZHANG Z X, XU W, WU H, et al. Identification of a potent enzyme for the detoxification of zearalenone[J]. Journal of Agricultural and Food Chemistry, 2020, 68(1):376-383.
[41] HU J Q, WANG G, HOU M X, et al. New hydrolase from Aeromicrobium sp. HA for the biodegradation of zearalenone: Identification, mechanism, and application[J]. Journal of Agricultural and Food Chemistry, 2023, 71(5):2411-2420.
[42] YU X R, TU T, LUO H Y, et al. Biochemical characterization and mutational analysis of a lactone hydrolase from Phialophora americana[J]. Journal of Agricultural and Food Chemistry, 2020, 68(8):2570-2577.
[43] JIANG T Z, WANG M X, LI X Y, et al. The replacement of main cap domain to improve the activity of a ZEN lactone hydrolase with broad substrate spectrum[J]. Biochemical Engineering Journal, 2022, 182:108418.
[44] WANG H, LU Z H, LIN X F, et al. The N-terminal hydrophobicity modulates a distal structural domain conformation of zearalenone lacton hydrolase and its application in protein engineering[J]. Enzyme and Microbial Technology, 2023, 165:110195.
[45] BI K, ZHANG W, XIAO Z Z, et al. Characterization, expression and application of a zearalenone degrading enzyme from Neurospora crassa[J]. AMB Express, 2018, 8(1):194.
[46] ZHANG Y, LIU X M, ZHANG Y P, et al. Cloning and characterization of three novel enzymes responsible for the detoxification of zearalenone[J]. Toxins, 2022, 14(2):82.
[47] YU Y S, WU H, TANG Y Q, et al. Cloning, expression of a peroxiredoxin gene from Acinetobacter sp. SM04 and characterization of its recombinant protein for zearalenone detoxification[J]. Microbiological Research, 2012, 167(3):121-126.
[48] GUO Y P, WANG Y N, TANG Y, et al. Combined in silico investigation and in vitro characterization of the zearalenone detoxification potential of dye-decolorizing peroxidase from Bacillus subtilis 168[J]. Food Control, 2023, 146:109549.
[49] QIN X, XIN Y Z, SU X Y, et al. Efficient degradation of zearalenone by dye-decolorizing peroxidase from Streptomyces thermocarboxydus combining catalytic properties of manganese peroxidase and laccase[J]. Toxins, 2021, 13(9):602.
[50] SUN F, YU D Z, ZHOU H Y, et al. CotA laccase from Bacillus licheniformis ZOM-1 effectively degrades zearalenone, aflatoxin B1 and alternariol[J]. Food Control, 2023, 145:109472.
[51] GUO Y P, WANG Y N, LIU Y R, et al. Detoxification of the mycoestrogen zearalenone by Bacillus licheniformis spore CotA laccase and application of immobilized laccase in contaminated corn meal[J]. LWT, 2022, 163:113548.
[52] QIN X, XIN Y Z, ZOU J H, et al. Efficient degradation of aflatoxin B1 and zearalenone by laccase-like multicopper oxidase from Streptomyces thermocarboxydus in the presence of mediators[J]. Toxins, 2021, 13(11):754.
[53] WANG X L, BAI Y G, HUANG H Q, et al. Degradation of aflatoxin B1 and zearalenone by bacterial and fungal laccases in presence of structurally defined chemicals and complex natural mediators[J]. Toxins, 2019, 11(10):609.
[54] TANG Y Q, XIAO J M, CHEN Y, et al. Secretory expression and characterization of a novel peroxiredoxin for zearalenone detoxification in Saccharomyces cerevisiae[J]. Microbiological Research, 2013, 168(1):6-11.
[55] MICHLMAYR H, VARGA E, LUPI F, et al. Synthesis of mono- and di-glucosides of zearalenone and α-/ β-Zearalenol by recombinant barley glucosyltransferase HvUGT14077[J]. Toxins, 2017, 9(2):58.
[56] PAN Y Y, LIU C D, YANG J G, et al. Conversion of zearalenone to β-Zearalenol and zearalenone-14, 16-diglucoside by Candida parapsilosis ATCC 7330[J]. Food Control, 2022, 131:108429.
[57] ZHU Y, DROUIN P, LEPP D, et al. A novel microbial zearalenone transformation through phosphorylation[J]. Toxins, 2021, 13(5):294.
[58] YANG S B, ZHENG H C, XU J Y, et al. New biotransformation mode of zearalenone identified in Bacillus subtilis Y816 revealing a novel ZEN conjugate[J]. Journal of Agricultural and Food Chemistry, 2021, 69(26):7409-7419.
[59] 赵程程, 孙长坡, 孙晶, 等. 一株新型降解玉米赤霉烯酮的沙福芽孢杆菌及其关键酶分析[J]. 食品科学, 2023, 44(22):165-172. ZHAO C C, SUN C P, SUN J, et al. Identification and characterization of a novel Bacillus safensis M7L4 with zearalenone removal capacity and its potent enzyme[J]. Food Science, 2023, 44(22):165-172.
[60] XU L P, SUN X L, WAN X H, et al. Identification of a Bacillus amyloliquefaciens H6 thioesterase involved in zearalenone detoxification by transcriptomic analysis[J]. Journal of Agricultural and Food Chemistry, 2020, 68(37):10071-10080.
[61] SUN X L, HE X X, XUE K S, et al. Biological detoxification of zearalenone by Aspergillus niger strain FS10[J]. Food and Chemical Toxicology, 2014, 72:76-82.
[62] JI J, YU J, XU W, et al. Isolation and mechanistic characterization of a novel zearalenone-degrading enzyme[J]. Foods, 2022, 11(18):2908.
[63] HE M L, LI Y, PI F W, et al. A novel detoxifying agent: Using rice husk carriers to immobilize zearalenone-degrading enzyme from Aspergillus niger FS10[J]. Food Control, 2016, 68:271-279.
[64] TIAN Y, TAN Y L, YAN Z, et al. Antagonistic and detoxification potentials of Trichoderma isolates for control of zearalenone (ZEN) producing Fusarium graminearum[J]. Frontiers in Microbiology, 2018, 8:2710.
[65] KRISZT R, KRIFATON C, SZOBOSZLAY S, et al. A new zearalenone biodegradation strategy using non-pathogenic Rhodococcus pyridinivorans K408 strain[J]. PLoS One, 2012, 7(9): e43608.
[66] VEKIRU E, HAMETNER C, MITTERBAUER R, et al. Cleavage of zearalenone by Trichosporon mycotoxinivorans to a novel nonestrogenic metabolite[J]. Applied and Environmental Microbiology, 2010, 76(7):2353-2359.
[67] TAN H, ZHANG Z M, HU Y C, et al. Isolation and characterization of Pseudomonas otitidis TH-N1 capable of degrading zearalenone[J]. Food Control, 2015, 47:285-290.
[68] TAN H, HU Y C, HE J, et al. Zearalenone degradation by two Pseudomonas strains from soil[J]. Mycotoxin Research, 2014, 30(4):191-196.
[69] GAO R Y, ZHENG J, LU W, et al. Shinella oryzae sp. nov., a novel zearalenone-resistant bacterium isolated from rice paddy soil[J]. Antonie Van Leeuwenhoek, 2022, 115(5):573-587.
[70] XU J H, WANG H J, ZHU Z W, et al. Isolation and characterization of Bacillus amyloliquefaciens ZDS-1: Exploring the degradation of zearalenone by Bacillus spp.[J]. Food Control, 2016, 68:244-250.
[71] LEI Y P, ZHAO L H, MA Q G, et al. Degradation of zearalenone in swine feed and feed ingredients by Bacillus subtilis ANSB01G[J]. World Mycotoxin Journal, 2014, 7(2):143-151.
[72] ZHAO L H, LEI Y P, BAO Y H, et al. Ameliorative effects of Bacillus subtilis ANSB01G on zearalenone toxicosis in pre-pubertal female gilts[J]. Food Additives &Contaminants. Part A, Chemistry, Analysis, Control, Exposure &Risk Assessment, 2015, 32(4):617-625.
[73] TINYIRO S E, WOKADALA C, XU D, et al. Adsorption and degradation of zearalenone by bacillus strains[J]. Folia Microbiologica, 2011, 56(4):321-327.
[74] 郝小龙, 常晓娇, 伍松陵, 等. 玉米赤霉烯酮降解酶ZLHY6活性影响因素研究[J]. 粮油食品科技, 2013, 21(3):99-101.HAO X L, CHANG X J, WU S L, et al. Study on the factors to affect the activity of zearalenone degrading enzyme ZLHY6[J]. Science and Technology of Cereals, Oils and Foods, 2013, 21(3):99-101.
[75] XU Z X, LIU W D, CHEN C C, et al. Enhanced α-Zearalenol hydrolyzing activity of a mycoestrogen-detoxifying lactonase by structure-based engineering[J]. ACS Catalysis, 2016, 6(11):7657-7663.
[76] 陈权, 吕成, 许菲. 基于柔性区域的计算设计改造玉米赤霉烯酮水解酶热稳定性[J]. 生物工程学报, 2021, 37(12):4415-4429.CHEN Q, LYU C, XU F. Computation-aided design of the flexible region of zearalenone hydrolase improves its thermal stability[J]. Chinese Journal of Biotechnology, 2021, 37(12):4415-4429.
[77] 官嫒林, 张萌, 许菲. 结构指导的玉米赤霉烯酮水解酶的热稳定性改造[J]. 生物工程学报, 2023, 39(8):3336-3350.GUAN A L, ZHANG M, XU F. Structure-guided engineering for improving the thermal stability of zearalenone hydrolase[J]. Chinese Journal of Biotechnology, 2023, 39(8):3336-3350.
[78] LIN M, TAN J, XU Z B, et al. Computational design of enhanced detoxification activity of a zearalenone lactonase from Clonostachys rosea in acidic medium[J]. RSC Advances, 2019, 9(54):31284-31295.
[79] 许中霞, 刘桂智, 刘卫东, 等. 玉米赤霉烯酮水解酶耐热性的分子改造[J]. 食品与生物技术学报, 2019, 38(7):71-77.XU Z X, LIU G Z, LIU W D, et al. Molecular engineering of mycoestrogen-detoxifying lactonase ZHD101 to improve enzyme thermostability[J]. Journal of Food Science and Biotechnology, 2019, 38(7):71-77.
[80] 杜昊澜, 骆树娟, Tosin Victor Adegoke, 等. 基于热不稳定区段分析和二硫键预测的玉米赤霉烯酮脱毒酶ZLHY-6热稳定性改造[J]. 食品安全质量检测学报, 2022, 13(22):7150-7157.DU H L, LUO S J, ADEGOKE T, et al. Thermal stability modification of zearalenone detoxification enzyme ZLHY-6 based on thermolabile segment analysis and disulfide bond prediction[J]. Journal of Food Safety &Quality, 2022, 13(22):7150-7157.
[81] FANG Y Y, HUANG Z L, XU W, et al. Efficient elimination of zearalenone at high processing temperatures by a robust mutant of Gliocladium roseum zearalenone lactonase[J]. Food Control, 2022, 142:109222.
[82] PENG W, KO T P, YANG Y Y, et al. Crystal structure and substrate-binding mode of the mycoestrogen-detoxifying lactonase ZHD from Clonostachys rosea[J]. RSC Advances, 2014, 4(107):62321-62325.
[83] LIU Y, WAN Y Z, ZHU J X, et al. Theoretical study on Zearalenol compounds binding with wild type zearalenone hydrolase and V153H mutant[J]. International Journal of Molecular Sciences, 2018, 19(9):2808.