百合(Lilium spp.)是百合科(Liliaceae)百合属(Lilium L.)植物的统称,多年生草本植物,地下部分为肉质鳞茎,其花姿绰约、花色多彩、气味芬芳,是世界上最重要的观赏花卉之一[1]。全世界已发现百合属约110种[2],中国作为百合产地之一,野生百合约47种[3]。百合不仅作为观赏花卉,其地下鳞茎部分也具有多种营养成分,包括多糖、蛋白、皂苷和膳食纤维等[4],具有抗氧化及抗衰老等作用[5]。兰州百合(Lilium davidii var.unicolor),鳞片大、肉质饱满、味道甘甜、口感好,是我国主要的食用百合之一,近些年来受广大消费者的喜欢[6]。兰州百合鳞茎的颜色出土时为白色,但在贮藏运输过程中,许多鳞片由于花青素的积累而变成紫红色,极大地降低了百合的商用价值[7]。
国内外针对于百合花青素的研究,大多集中在花瓣部分,对于百合鳞茎花青素的研究非常少。百合中花青素的主要成分为矢车菊素3-O-β-芸香糖苷,其次是3-O-β-芸香糖-7-O-β-葡萄糖苷[8]。影响百合花青素生物合成途径的相关基因分为结构基因与调控基因。亚洲百合杂交品种‘Montreux’中,鉴定出查尔酮合成酶(chalcone synthase,CHS)与二氢黄酮醇还原酶(dihydroflavonol reductase,DFR)基因在百合花青素生物合成过程中发挥关键作用[9]。在东方百合‘Sorbonne’中,发现查尔酮异构酶(chalcone isomerase,CHI)和DFR基因与花青素合成紧密相关[10-11]。亚洲杂交百合‘Montreux’中,分离出2个MYB(v-myb avian myeloblastosis viral oncogene homolog)基因:LhMYB6与LhMYB12,参与花青素的生物合成[12]。岷江百合(Lilium.regale)中,LrMYB15在外侧花被、叶和苞片上游不同程度的表达,决定了花青素的合成与积累[13]。亚洲百合杂交品种‘Montreux’中,还鉴定有2个bHLH(basic helix-loop-helix)基因:LhbHLH1和LhbHLH2参与了花青素的合成且关系密切[14]。岷江百合(Lilium.regale)中发现,在LrbHLH2与LrMYB15协同作用下调节花青素的合成与积累[13]。
当前对于百合鳞茎花青素的研究较少,且针对百合花青素转录因子的研究多是以单个基因的克隆与分析为主,缺乏针对转录因子家族的鉴定与分析。本研究基于兰州百合鳞茎测序的转录组学数据鉴定出的MYB与bHLH转录因子家族进行筛选分析,并筛选出一些花青素相关的转录因子基因作为候选基因,分析结果将为深入研究兰州百合鳞茎中MYB与bHLH转录因子参与花青素生物合成的机制提供基础理论依据。
1 材料与方法
1.1 植物材料
百合鳞茎样本选自甘肃省兰州市西果园街道,大小均匀,无病虫害,无机械损伤。取百合鳞片第四层和第五层,用蒸馏水洗涤,浸于1 g/L次氯酸钠水溶液中浸泡1 min,消除潜在微生物,作为实验处理的植物材料[15]。将鳞片密封在聚乙烯薄膜塑料袋中(200 mm×150 mm,0.03 mm厚)。然后将鳞茎置于人工气候培养箱中,(20±2) ℃处理5 d(日照12 h,遮阳12 h),相对湿度保持在70%~80%。分别于第0天和第5天取百合鳞茎,用液氮快速冷冻,然后收集在5 mL的低温保存管中,送至上海百趣生物科技有限公司测转录组。每个实验设置3次生物学重复。
1.2 兰州百合鳞茎MYB与bHLH转录因子家族的理化性质分析、亚细胞定位预测
利用在线ProtParam网站(https://web.expasy.org/protparam/)对转录组鉴定出的MYB和bHLH类转录因子蛋白性质进行理化性质分析[16]。用在线WOLF PSORT网站(https://wolfpsort.hgc.jp/)对候选MYB与bHLH基因编码的蛋白进行亚细胞定位预测分析。然后参考DUBOS等[17]的方法,将转录组中MYB分类。
1.3 兰州百合鳞茎与拟南芥中MYB与bHLH转录因子进化关系、功能分析及候选转录因子的保守结构域分析
在拟南芥信息数据库TAIR(http://www.arabidopsis.org)获取拟南芥MYB与bHLH转录因子氨基酸序列。使用MEGA X软件分别构建兰州百合鳞茎MYB转录因子与拟南芥MYB转录因子家族系统发育树,及兰州百合鳞茎bHLH转录因子与拟南芥bHLH转录因子家族系统发育树,选用邻接(neighbor-joining, NJ)法对序列结果进行比对,选用Poisson模型并将Bootstrap value设置为1 000,缺失值处理方式为配对删除(pairwise deletion),其他参数使用默认值[18]。利用DNAMANV 6软件对兰州百合鳞MYB与bHLH转录因子进行多序列比对,并通过CDD v3.17(https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi/)与Pfamv32.0(https://pfam.xfam.org/)分析候选MYB与bHLH转录因子的保守结构域。
2 结果与分析
在兰州百合鳞茎转录组测序数据中,初步筛选到53个MYB转录因子相关序列,73个bHLH转录因子相关序列,运用NCBI在线blastp软件进行转录因子氨基酸序列比对筛选,最终鉴定出MYB转录因子50个,bHLH转录因子73个。
2.1 兰州百合鳞茎MYB转录因子
2.1.1 兰州百合鳞茎MYB转录因子的理化性质分析、亚细胞定位预测
针对鉴定出的兰州百合鳞茎MYB转录因子理化性质进行分析,其中包括蛋白长度、分子质量、等电点和亲疏水性。结果如表1所示,兰州百合鳞茎转录组中筛选出的50个MYB转录因子蛋白长度为93~1 365 aa,分子质量大小为10.93~152.08 kDa,等电点为4.63~11.63。预测结果还显示出50个MYB转录因子编码的蛋白不稳定指数均大于40,为不稳定蛋白,且50个蛋白的平均亲水指数均小于0,属于亲水性蛋白。亚细胞定位预测结果显示Cluster-96.0蛋白定位于线粒体,Cluster-4138.2991蛋白定位于细胞质,其余48个MYB蛋白定位于细胞核中。将兰州百合鳞茎转录组中50个MYB蛋白序列逐个进行在线BLAST,MYB分类结果如表1所示,2个1R-MYB类,44个R2R3-MYB类,3个3R-MYB类,1个4R-MYB类。兰州百合鳞茎转录组数据中MYB分类结果表示,1R-MYB占4%,R2R3-MYB占88%,3R-MYB占6%,4R-MYB占2%,不同类别的MYB转录因子分布比例与小麦(Triticum aestivum)、杨树(Populus spp.)、谷子(Setaria italica)和黑果枸杞(Lycium ruthenicum)等研究结果类似[17]。
2.1.2 兰州百合鳞茎与拟南芥中MYB转录因子进化关系、功能分析及候选转录因子的保守结构域分析
利用MEGA X软件对兰州百合鳞茎MYB转录因子与注释明确的拟南芥MYB转录因子蛋白家族构建系统进化树,分析进化关系与功能预测。根据DUBOS等[17]针对拟南芥MYB转录因子亚族分类,图1中使用红色显著标出的AtMYB75、AtMYB90、AtMYB113和AtMYB114属于S6亚组,拟南芥中功能明确的调控花青素生物合成晚期基因并促进花青素积累的转录因子[19]。拟南芥AtMYB75和AtMYB90对于DFR基因的表达至关重要[20],AtMYB113和AtMYB114影响花青素生物合成后期酶的表达[21]。图1中Cluster-668.0、Cluster-4138.14450、Cluster-4138.3405、Cluster-4138.19859和Cluster-4138.19860与S6亚组聚集在一起,推测其具有相似的花青素生物合成相关功能。
表1 兰州百合鳞茎MYB转录因子的理化性质以及亚细胞定位预测
Table 1 Physicochemical properties and prediction of subcellular localization of MYB transcription factors in Lilium davidii var.unicolor bulbs

基因IDMYB类型蛋白长度/aa分子质量/Da等电点平均亲水指数不稳定指数亚细胞定位Cluster-4138.15597R2R3-MYB13114 787.7910.28-0.64473.74细胞核Cluster-4138.15951R2R3-MYB24328 033.9111.63-0.73471.48细胞核Cluster-18210.0R2R3-MYB13716 107.159.39-0.99362.7细胞核Cluster-4138.12365R2R3-MYB40342 420.149.19-0.21278.49细胞核Cluster-4138.13270R2R3-MYB35240 670.49.3-0.88371.85细胞核Cluster-4138.188923R-MYB39944 875.797.25-0.94959.95细胞核Cluster-4138.3299R2R3-MYB29633 205.996.63-0.71864.93细胞核Cluster-4138.59343R-MYB45651 351.897.27-0.7271.03细胞核Cluster-4138.6563R2R3-MYB25929 179.098.74-0.66860.66细胞核Cluster-4138.663R2R3-MYB29331 657.35.73-0.4761.37细胞核Cluster-96.0R2R3-MYB10312 248.1810.28-0.96663.59线粒体Cluster-14547.03R-MYB67775 827.299.05-0.75569.11细胞核Cluster-16981.0R2R3-MYB47052 235.15.62-0.71764.96细胞核Cluster-17373.0R2R3-MYB20823 767.728.69-0.75169.9细胞核Cluster-18565.0R2R3-MYB26028 6237.09-0.67669.88细胞核Cluster-4138.15587R2R3-MYB23726 829.26.08-0.61473.25细胞核Cluster-4138.15589R2R3-MYB9310 932.699.2-0.60981.83细胞核Cluster-4138.4469R2R3-MYB56661 217.945.16-0.58561.38细胞核Cluster-4138.7249R2R3-MYB52258 127.855.82-0.67370.08细胞核Cluster-22293.0R2R3-MYB19422 749.799.24-1.08457.78细胞核Cluster-4138.12910R2R3-MYB1365152 086.344.7-0.81174.11细胞核Cluster-4138.13884R2R3-MYB67676 503.338.19-0.83969.16细胞核Cluster-4138.710R2R3-MYB9711 390.2410.12-1.07667.42细胞核Cluster-20279.0R2R3-MYB37642 486.416.83-0.80560.19细胞核Cluster-22102.0R2R3-MYB22024 559.715.39-0.5881.18细胞核Cluster-4138.15532R2R3-MYB32435 307.456.02-0.53771.98细胞核Cluster-4138.35484R-MYB92110 2762.048.8-0.65770.13细胞核Cluster-4138.4518R2R3-MYB26230 776.0511.23-1.00261.03细胞核Cluster-12765.0R2R3-MYB23727 222.546.34-0.66871.14细胞核Cluster-4138.14450R2R3-MYB24628 274.928.5-0.47391.5细胞核Cluster-4138.34051R-MYB16919 269.77.89-0.70476.75细胞核Cluster-4138.4351R2R3-MYB29833 545.796.32-0.66484.06细胞核Cluster-24169.0R2R3-MYB23626 669.19.51-0.66165.42细胞核Cluster-4138.22530R2R3-MYB33137 879.056.01-0.45689.27细胞核Cluster-4138.7808R2R3-MYB28931 778.18.69-0.55676.96细胞核Cluster-889.01R-MYB37741 573.544.87-0.68665.7细胞核Cluster-10852.0R2R3-MYB34938 445.955.21-0.69464.13细胞核Cluster-13894.0R2R3-MYB22525 544.088.63-0.68773.69细胞核Cluster-18761.0R2R3-MYB23426 185.838.82-0.54580.85细胞核Cluster-4138.1679R2R3-MYB34238 687.795.31-0.41685.53细胞核Cluster-4138.767R2R3-MYB25928 752.528.33-0.53779.58细胞核Cluster-668.0R2R3-MYB21724 919.679.42-0.75383.59细胞核Cluster-4138.13060R2R3-MYB22825 717.277.72-0.59577.06细胞核Cluster-4138.19859R2R3-MYB11513 213.3110.18-0.51594.09细胞核Cluster-4138.19860R2R3-MYB23026 344.028.91-0.6983.52细胞核Cluster-1401.0R2R3-MYB16118 674.958.74-1.01956.34细胞核Cluster-1443.0R2R3-MYB24627 727.067.25-0.59278.9细胞核Cluster-4138.2317R2R3-MYB30033 928.885.22-0.68674.8细胞核Cluster-4138.2991R2R3-MYB29531 658.716.28-0.52763.86细胞质Cluster-868.0R2R3-MYB20822 286.688.83-0.84165.82细胞核

图1 兰州百合鳞茎MYB转录因子与拟南芥AtMYB转录因子进化分析
Fig.1 Phylogenetic tree of MYB transcription factor in Lilium davidii var.unicolor bulbs and Arabidopsis thaliana AtMYB transcription factor
将上述5个兰州百合鳞茎MYB转录因子作为候选MYB转录因子,并利用在线软件进行蛋白序列保守结构域的预测。预测结果如图2显示,5个候选MYB转录因子中Cluster-4138.3405仅含有1个 高度保守MYB-binding结构域,属于1R-MYB转录因子,其余4个经预测均含有2个高度保守MYB-binding结构域,属于R2R3-MYB转录因子。该结果与拟南芥MYB转录因子的研究中MYB转录因子N端存在高度保守DNA-binding结合结构域的结果类似[21]。蛋白保守结构域分析预测还发现MYB-binding结构域的蛋白数量为30~50个氨基酸,该结果与绢毛委陵菜(Potentilla sericea)[22]、漆树(Toxicodendron vernicifluum)[23]和亚洲百合(Lilium Asiatic hybrids ‘Little Kiss’)[24]MYB的研究类似。
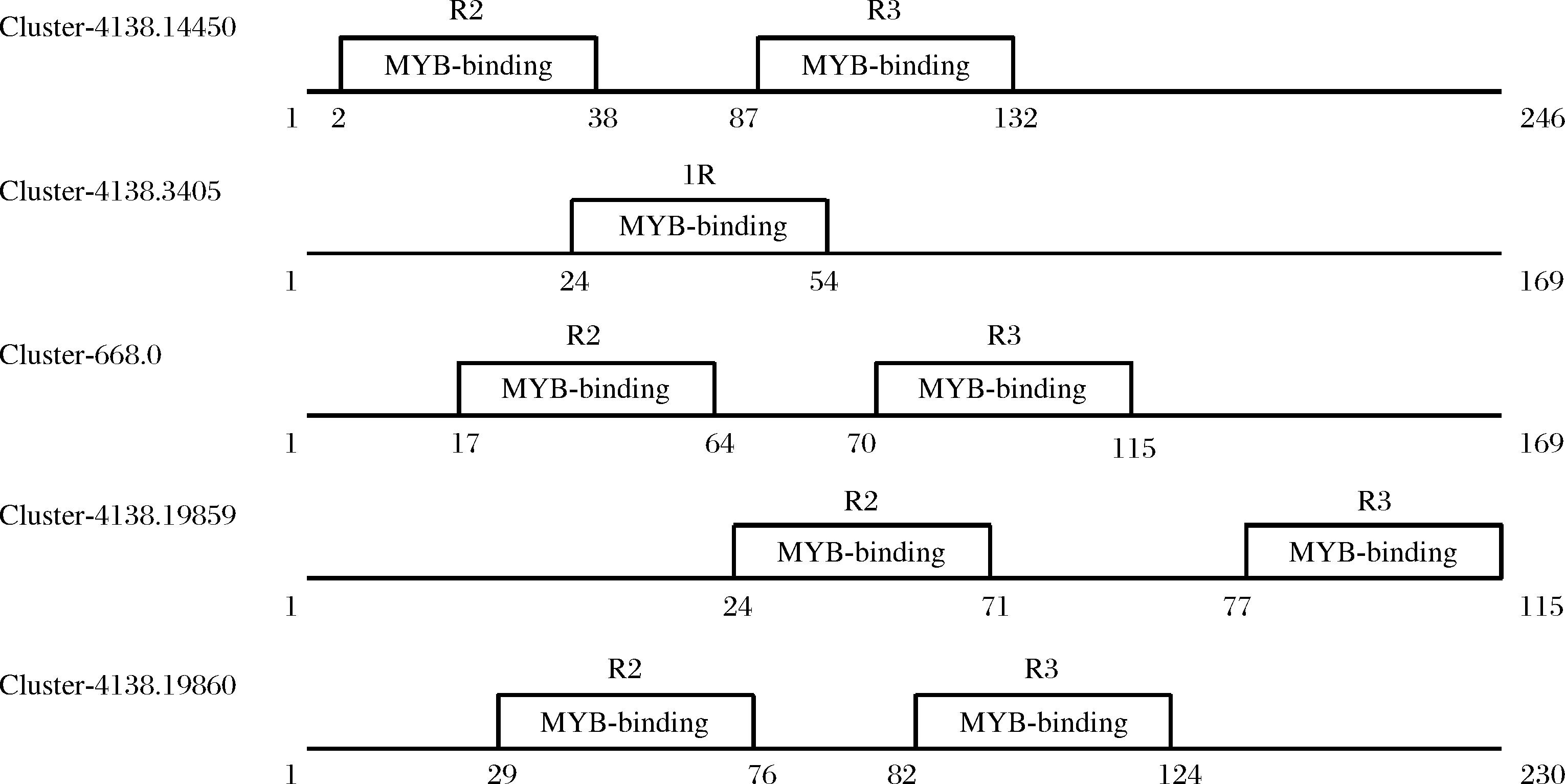
图2 兰州百合鳞茎花青素相关MYB转录因子蛋白保守结构预测
Fig.2 Prediction of the conserved structure of anthocyanin-related MYB transcription factor proteins in Lilium davidii var.unicolor bulbs
2.2 兰州百合鳞茎bHLH转录因子
2.2.1 兰州百合鳞茎bHLH转录因子的理化性质分析、亚细胞定位预测
对鉴定出的73个兰州百合鳞茎bHLH转录因子进行理化性质分析。结果如表2所示,73个bHLH转录因子的蛋白长度为39~703 aa,分子质量为4.61~79.85 kDa,等电点为3.8~11.47。在线预测结果表明73个兰州百合鳞茎bHLH转录因子编码的蛋白均为不稳定蛋白(不稳定指数大于40),并且仅有一个Cluster-4138.21820平均亲水指数大于0为疏水性蛋白,另外72个bHLH编码蛋白均为亲水性蛋白。亚细胞定位预测结果表明,1个bHLH定位于细胞外,58个定位于细胞核,8个定位于叶绿体,2个定位于线粒体,1个定位于内质网,3个定位于细胞质。转录组中73个bHLH经亚细胞定位,在细胞各位置中的分布比例类似于银杏(Ginkgo)[25]与黑果枸杞(Lycium ruthenicum Murr.)[26]。
表2 兰州百合鳞茎bHLH转录因子的理化性质以及亚细胞定位预测
Table 2 Physicochemical properties and prediction of subcellular localization of bHLH transcription factors in Lilium davidii var.unicolor bulbs

基因ID蛋白长度/aa分子质量/Da等电点平均亲水指数不稳定指数亚细胞定位Cluster-18294.069678 007.95.02-0.54379.12细胞核Cluster-25735.017620 212.649.74-0.207107.33叶绿体Cluster-25784.037741 928.426.79-0.60664.48细胞核Cluster-4138.1112357161 521.495.35-0.52663.15细胞核Cluster-4138.1132726129 803.976.21-0.37485.44细胞核Cluster-4138.1151916919 3107.77-0.53688.22细胞核Cluster-4138.1313443147 567.85.94-0.81561.6细胞核Cluster-4138.159679711 025.528.5-0.231106.39细胞质Cluster-4138.1916935839 114.855.04-0.71267.54细胞核Cluster-4138.2182026829 601.85.770.147101.87内质网Cluster-4138.21825728 411.5511.47-1.39459.58细胞核Cluster-4138.635454258 656.656.01-0.68663.75细胞核Cluster-905.044549 033.748.08-0.63570.58细胞核Cluster-16182.030834 069.734.34-0.3778.15细胞核Cluster-16511.036540 919.775.15-0.54377.29细胞核Cluster-17568.025528 180.917.29-0.62473.45细胞核Cluster-20882.028331 385.186.28-0.57176.57细胞核Cluster-21617.033737 037.455.96-0.6859.11细胞核Cluster-23591.012114 035.457.96-1.07661.32叶绿体Cluster-4138.12406515 768.647.03-1.04567.06细胞核Cluster-4138.1469125228 212.369.25-0.25783.93细胞核Cluster-4138.1662827229 663.45.44-0.6762.98细胞核Cluster-4138.17652859 791.664.62-0.91867.65细胞核Cluster-4138.1870122925 029.38.42-0.36967.29细胞核Cluster-4138.383831535 554.086.12-0.90560.95细胞核Cluster-4138.677126629 529.545.46-0.64463.5细胞核Cluster-4138.864833637 078.017.31-0.38287.11细胞核Cluster-4138.926514617 036.864.95-0.51262.05细胞核Cluster-4138.981420322 514.079.59-0.4387.93细胞核Cluster-21537.038342 847.918.96-0.40685.59细胞核
续表2
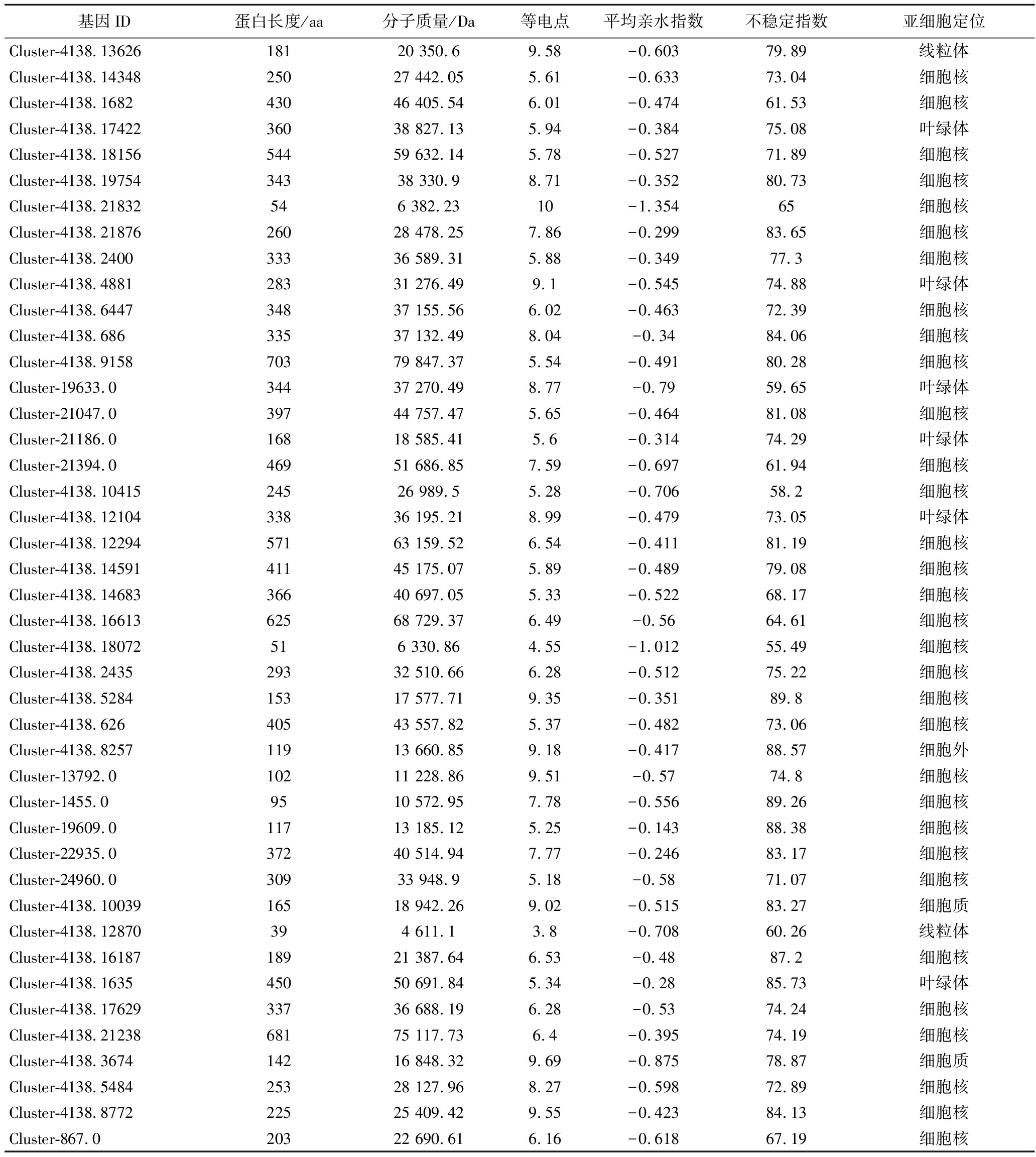
基因ID蛋白长度/aa分子质量/Da等电点平均亲水指数不稳定指数亚细胞定位Cluster-4138.1362618120 350.69.58-0.60379.89线粒体Cluster-4138.1434825027 442.055.61-0.63373.04细胞核Cluster-4138.168243046 405.546.01-0.47461.53细胞核Cluster-4138.1742236038 827.135.94-0.38475.08叶绿体Cluster-4138.1815654459 632.145.78-0.52771.89细胞核Cluster-4138.1975434338 330.98.71-0.35280.73细胞核Cluster-4138.21832546 382.2310-1.35465细胞核Cluster-4138.2187626028 478.257.86-0.29983.65细胞核Cluster-4138.240033336 589.315.88-0.34977.3细胞核Cluster-4138.488128331 276.499.1-0.54574.88叶绿体Cluster-4138.644734837 155.566.02-0.46372.39细胞核Cluster-4138.68633537 132.498.04-0.3484.06细胞核Cluster-4138.915870379 847.375.54-0.49180.28细胞核Cluster-19633.034437 270.498.77-0.7959.65叶绿体Cluster-21047.039744 757.475.65-0.46481.08细胞核Cluster-21186.016818 585.415.6-0.31474.29叶绿体Cluster-21394.046951 686.857.59-0.69761.94细胞核Cluster-4138.1041524526 989.55.28-0.70658.2细胞核Cluster-4138.1210433836 195.218.99-0.47973.05叶绿体Cluster-4138.1229457163 159.526.54-0.41181.19细胞核Cluster-4138.1459141145 175.075.89-0.48979.08细胞核Cluster-4138.1468336640 697.055.33-0.52268.17细胞核Cluster-4138.1661362568 729.376.49-0.5664.61细胞核Cluster-4138.18072516 330.864.55-1.01255.49细胞核Cluster-4138.243529332 510.666.28-0.51275.22细胞核Cluster-4138.528415317 577.719.35-0.35189.8细胞核Cluster-4138.62640543 557.825.37-0.48273.06细胞核Cluster-4138.825711913 660.859.18-0.41788.57细胞外Cluster-13792.010211 228.869.51-0.5774.8细胞核Cluster-1455.09510 572.957.78-0.55689.26细胞核Cluster-19609.011713 185.125.25-0.14388.38细胞核Cluster-22935.037240 514.947.77-0.24683.17细胞核Cluster-24960.030933 948.95.18-0.5871.07细胞核Cluster-4138.1003916518 942.269.02-0.51583.27细胞质Cluster-4138.12870394 611.13.8-0.70860.26线粒体Cluster-4138.1618718921 387.646.53-0.4887.2细胞核Cluster-4138.163545050 691.845.34-0.2885.73叶绿体Cluster-4138.1762933736 688.196.28-0.5374.24细胞核Cluster-4138.2123868175 117.736.4-0.39574.19细胞核Cluster-4138.367414216 848.329.69-0.87578.87细胞质Cluster-4138.548425328 127.968.27-0.59872.89细胞核Cluster-4138.877222525 409.429.55-0.42384.13细胞核Cluster-867.020322 690.616.16-0.61867.19细胞核
2.2.2 兰州百合鳞茎与拟南芥中bHLH转录因子进化关系、功能分析及候选转录因子的保守结构域分析
利用MEGAX软件对兰州百合鳞茎bHLH转录因子与注释明确的拟南芥bHLH转录因子蛋白家族构建系统进化树,分析进化关系与功能预测。根据拟南芥bHLH亚族分类,图3中使用红色显著标出的分支AtbHLH42、AtbHLH12、AtbHLH1和AtbHLH2属于IIIf亚族是拟南芥中功能明确的花青素生物合成相关的转录因子[27],AtbHLH32可作为花青素的负调节剂影响拟南芥花青素的合成[28],AtbHLH42可与AtMYB75协同作用通过影响DFR基因的表达,进而影响花青素的生物合成[29]。其中Cluster-4138.9158、Cluster-18294.0与IIIf亚族聚集在一起,推测其可能具有较高的亲缘性以及相似的花青素生物合成相关功能。
将上述2个兰州百合鳞茎bHLH转录因子作为候选bHLH转录因子,并进行蛋白序列保守结构域的预测。预测结果如图4显示,2个候选bHLH转录因子均在C端预测出螺旋-环-螺旋(helix-loop-helix,HLH)保守域,分别由50个和64个氨基酸构成,这一结果符合bHLH转录因子的基本特征,与谷子(Setaria italica L.)[30]和黑果枸杞[26]等bHLH研究结果类似。

图3 兰州百合鳞茎bHLH转录因子与拟南芥AtbHLH转录因子进化分析
Fig.3 Phylogenetic tree of bHLH transcription factor in Lilium davidii var.unicolor bulbs and Arabidopsis thaliana AtbHLH transcription factor

图4 兰州百合鳞茎花青素相关bHLH转录因子蛋白保守结构预测
Fig.4 Prediction of the conserved structure of anthocyanin-related bHLH transcription factor proteins in Lilium davidii var.unicolor bulbs
2.3 兰州百合鳞茎花青素相关转录因子基因表达分析
基于兰州百合鳞茎转录组数据,本研究对比了第0天和第5天的兰州百合鳞茎分析花青素相关转录因子基因表达量。如表3所示,第0天与第5天各基因的表达量来自于兰州百合鳞茎转录组数据。研究表明,2个候选花青素相关bHLH转录因子中,Cluster-18294.0在第5天的表达量略低于第0天,但P值为0.902 95,表示兰州百合鳞茎第0天VS第5天中该基因无显著性表达差异,而Cluster-4138.9158表达量显示出明显的上升趋势。5个候选花青素相关MYB转录因子中,Cluster-413814450在第5天的表达量呈现下降趋势,且存在极其显著的表达差异,其余4个候选MYB转录因子在第5天皆呈上升趋势,并具有显著性差异。
表3 兰州百合鳞茎花青素相关转录因子基因的表达量分析
Table 3 Expression analysis of anthocyanin-related transcription factor genes in Lilium davidii var.unicolor bulbs
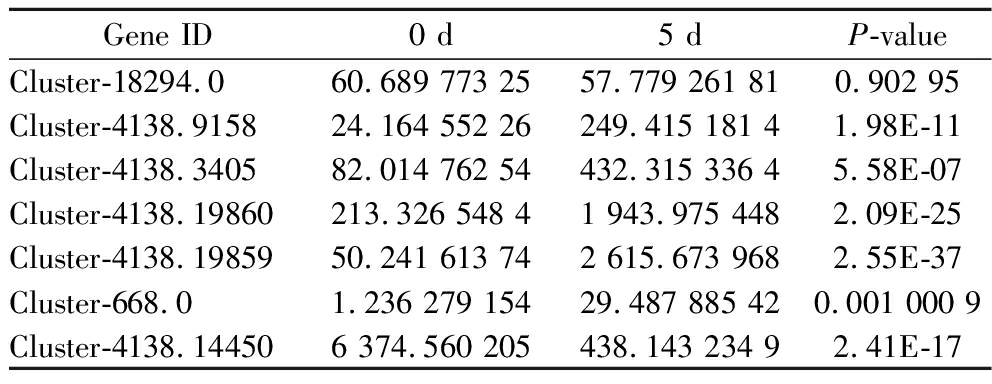
Gene ID0 d 5 d P-valueCluster-18294.060.689 773 2557.779 261 810.902 95Cluster-4138.915824.164 552 26249.415 181 41.98E-11Cluster-4138.340582.014 762 54432.315 336 45.58E-07Cluster-4138.19860213.326 548 41 943.975 4482.09E-25Cluster-4138.1985950.241 613 742 615.673 9682.55E-37Cluster-668.01.236 279 15429.487 885 420.001 000 9Cluster-4138.144506 374.560 205438.143 234 92.41E-17
注:Gene ID为基因编号,0 d为第0天兰州百合鳞茎基因表达量,5 d为第5天兰州百合鳞茎基因表达量,P-value为统计学差异显著性分析P值。
植物花青素合成过程中,调控基因通过编码转录因子调控结构基因的表达量,进而影响花青素的合成通路。本课题组关于兰州百合鳞茎的转录组与代谢组研究中[31],花青素合成通路中共发现20个差异表达结构基因。7个候选花青素相关转录因子基因中Cluster-18294.0未在第5天与第0天发现差异表达,故推测其在兰州百合鳞茎花青素合成中并不起调控作用。利用皮尔逊系数法测定候选基因中6个差异表达转录因子基因与20个差异表达结构基因的相关性。结果如图5所示,Cluster-4138.14450与19个高表达花青素相关结构基因呈现负相关关系,推测其在兰州百合鳞茎花青素积累过程中发挥抑制作用,Cluster-668.0与5个差异表达的结构基因显示出负相关,与其他15个差异表达结构基因显示出正相关关系,且皮尔逊系数普遍较低,故推测Cluster-668.0对于兰州百合鳞茎花青素合成的影响力较弱。其余4个候选花青素相关转录因子基因与19个高表达花青素相关结构基因均呈现正相关关系,且部分皮尔逊相关性系数大于0.8,呈现出极正相关关系。因此推测1个bHLH转录因子基因(Cluster-4138.9158),3个MYB转录因子基因(Cluster-4138.3405,Cluster-4138.19860与Cluster-4138.19859),对兰州百合鳞茎花青素的合成积累起促进作用。

图5 兰州百合鳞茎花青素相关转录因子基因与结构基因相关性分析
Fig.5 Correlation analysis of anthocyanin-related transcription factor genes and structural genes in Lilium davidii var.unicolor bulbs
3 结论
综上所述,本研究基于转录组数据筛选分析了兰州百合鳞茎的MYB与bHLH转录因子家族,并通过与模式植物拟南芥注释明确的MYB与bHLH转录因子构建系统进化树分析亲缘关系,以及利用皮尔逊系数分析花青素相关差异表达转录因子与结构基因的相关性,初步筛选出兰州百合鳞茎花青素相关的3个MYB基因(Cluster-4138.3405,Cluster-4138.19860与Cluster-4138.19859)和1个bHLH基因(Cluster-4138.9158)发挥促进作用,1个MYB基因(Cluster-4138.14450)起抑制作用,1个MYB基因(Cluster-668.0)影响力较弱,1个bHLH基因(Cluster-18294.0)未起调控作用。以上研究结果为进一步深入研究百合鳞茎花青素相关的分子调控机制提供理论基础。
[1] 陈俊愉. 中国花卉品种分类学[M].北京:中国林业出版社, 2001.
CHEN J Y.Taxonomy of Chinese Flower Varieties[M].Beijing:China Forestry Press, 2001.
[2] 夏婷, 耿兴敏, 罗凤霞.不同花色野生百合色素成分分析[J].东北林业大学学报, 2013, 41(5):109-113;166.
XIA T, GENG X M, LUO F X.Components of flower pigments in the petals of wild Lilium in China[J].Journal of Northeast Forestry University, 2013, 41(5):109-113;166.
[3] 王瑜, 崔金腾, 张克中, 等.百合花青素苷合成酶基因片段的克隆及表达分析[J].中国农学通报, 2013, 29(10):162-166.
WANG Y, CUI J T, ZHANG K Z, et al.Molecular cloning and expression analysis of anthocyanidin synthase gene fragment in Lilium[J].Chinese Agricultural Science Bulletin, 2013, 29(10):162-166.
[4] 胡悦, 杜运鹏, 田翠杰, 等.百合属植物化学成分及其生物活性的研究进展[J].食品科学, 2018, 39(15):323-332.
HU Y, DU Y P, TIAN C J, et al.A review of chemical components and their bioactivities from the genus Lilium[J].Food Science, 2018, 39(15):323-332.
[5] KANO M, TAKAYANAGI T, HARADA K, et al.Antioxidative activity of anthocyanins from purple sweet potato, Ipomoera batatas cultivar Ayamurasaki[J].Bioscience, Biotechnology, and Biochemistry, 2005, 69(5):979-988.
[6] 范国良, 刘养卉.兰州百合产业现状与研究进展[J].甘肃农业, 2013(4):12-13.
FAN G L, LIU Y H.Present situation and research progress of lily industry in Lanzhou[J].Gansu Agriculture, 2013(4):12-13.
[7] 肖伟. 兰州百合LdMYB6基因的克隆与初步功能分析[D].长春:吉林农业大学, 2019.
XIAO W.Cloning and preliminary functional analysis of LdMYB6 gene from Lilium davidii var. unicolor[D].Changchun:Jilin Agricultural University, 2019.
[8] NØRBAEK R, KONDO T.Anthocyanins from flowers of Lilium (Liliaceae)[J].Phytochemistry, 1999, 50(7):1181-1184.
[9] 陈洁, 安利清, 王涛, 等.百合查尔酮合成酶基因克隆及其转化烟草的花色表达分析[J].西北植物学报, 2012, 32(8):1511-1517. CHEN J, AN L Q, WANG T, et al.Cloning of chalcone synthase gene in Lilium and expression analysis of flower colour changes in transgenic tobacco[J].Acta Botanica Boreali-Occidentalia Sinica, 2012, 32(8):1511-1517.
[10] 覃仁娟. 百合DFR基因克隆及功能初步鉴定[D].雅安:四川农业大学, 2014.
QIN R J.Cloning and functional initial characterization of the gene encoding dihydroflavonol 4-reductase (DFR) from lily[D].Ya′an:Sichuan Agricultural University, 2014.
[11] 窦晓莹, 郎利新, 包放, 等.东方百合查尔酮异构酶基因LhCHI的克隆及表达[J].东北林业大学学报, 2015,43(9):6-11;17.
DOU X Y, LANG L X, BAO F, et al.Cloning and expression analysis of chalcone isomerase gene LhCHI in oriental hybrid lily (Lilium spp.)[J].Journal of Northeast Forestry University, 2015,43(9):6-11;17.
[12] YAMAGISHI M, SHIMOYAMADA Y, NAKATSUKA T, et al.Two R2R3-MYB genes, homologs of Petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of Asiatic hybrid lily[J].Plant and Cell Physiology, 2010, 51(3):463-474.
[13] YAMAGISHI M.A novel R2R3-MYB transcription factor regulates light-mediated floral and vegetative anthocyanin pigmentation patterns in Lilium regale[J].Molecular Breeding, 2016, 36(1):1-14.
[14] NAKATSUKA A, YAMAGISHI M, NAKANO M, et al.Light-induced expression of basic helix-loop-helix genes involved in anthocyanin biosynthesis in flowers and leaves of Asiatic hybrid lily[J].Scientia Horticulturae, 2009, 121(1):84-91.
[15] HUANG D J, LI W T, DAWUDA M M, et al.Hydrogen sulfide reduced colour change in Lanzhou lily-bulb scales[J].Postharvest Biology and Technology, 2021, 176:111520.
[16] GARG V K, AVASHTHI H, TIWARI A, et al.MFPPI-multi FASTA ProtParam interface[J].Bioinformation, 2016, 12(2):74-77.
[17] DUBOS C, STRACKE R, GROTEWOLD E, et al.MYB transcription factors in Arabidopsis[J].Trends in Plant Science, 2010, 15(10):573-581.
[18] THORLEY J L, PAGE R D M.RadCon:Phylogenetic tree comparison and consensus[J].Bioinformatics, 2000, 16(5):486-487.
[19] GONZALEZ A, ZHAO M Z, LEAVITT J M, et al.Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings[J].The Plant Journal, 2008, 53(5):814-827.
[20] KRANZ H D, DENEKAMP M, GRECO R, et al.Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana[J].The Plant Journal, 1998, 16(2):263-276.
[21] STRACKE R, WERBER M, WEISSHAAR B.The R2R3-MYB gene family in Arabidopsis thaliana[J].Current Opinion in Plant Biology, 2001, 4(5):447-456.
[22] 刘振, 武俊刚, 李响, 等.基于转录组数据的绢毛委陵菜MYB转录因子家族分析[J/OL].分子植物育种, 2021.http://kns.cnki.net/kcms/detail/46.1068.S.20210722.1457.024.html.
LIU Z, WU J G, LI X, et al.Identification of the MYB transcription factor family of Potentilla sericata and screening of the MYB gene regulating heavy metal Cadmium[J/OL].Molecular Plant Breeding, 2021.http://kns.cnki.net/kcms/detail/46.1068.S.20210722.1457.024.html.
[23] 谢冬冬, 王武萍, 何学高, 等.基于转录组的漆树MYB转录因子的筛选及分析[J].西北林学院学报, 2021, 36(1):108-116. XIE D D, WANG W P, HE X G, et al.Screening and analysis of MYB transcription factors based on transcriptome data in Toxicodendron vernicifluum[J].Journal of Northwest Forestry University, 2021, 36(1):108-116.
[24] 王雪倩, 袁国振, 吴泽, 等.亚洲百合MYB转录因子家族的鉴定及调控花粉败育MYB基因的筛选[J].农业生物技术学报, 2019, 27(11):1951-1961.
WANG X Q, YUAN G Z, WU Z, et al.Identification of MYB transcription factors family and screening MYB genes associated with pollen abortion in Lilium Asiatic hybrids[J].Journal of Agricultural Biotechnology, 2019, 27(11):1951-1961.
[25] 冯磊, 石元豹, 汪贵斌, 等.银杏bHLH家族转录因子生物信息学及表达分析[J].江苏农业学报, 2019, 35(2):400-411.
FENG L, SHI Y B, WANG G B, et al.Bioinformatics and expression analysis of transcription factors of ginkgo bHLH family[J].Jiangsu Journal of Agricultural Sciences, 2019, 35(2):400-411.
[26] 刘静, 王翠平, 朱强, 等.黑果枸杞bHLH转录因子家族的生物信息学分析[J].分子植物育种, 2020, 18(14):4612-4623.
LIU J, WANG C P, ZHU Q, et al.Bioinformatics analysis of bHLH transcription factor family in Lycium ruthenicum Murr.[J].Molecular Plant Breeding, 2020, 18(14):4612-4623.
[27] HEIM M A, JAKOBY M, WERBER M, et al.The basic helix-loop-helix transcription factor family in plants:A genome-wide study of protein structure and functional diversity[J].Molecular Biology and Evolution, 2003, 20(5):735-747.
[28] SATO S, NAKAMURA Y, KANEKO T, et al.Structural analysis of Arabidopsis thaliana chromosome 3.I.sequence features of the regions of 4,504,864 bp covered by sixty P1 and TAC clones[J].DNA Research, 2000, 7(2):131-135.
[29] NESI N, DEBEAUJON I, JOND C, et al.The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques[J].The Plant Cell, 2000, 12(10):1863-1878.
[30] 孙颖琦, 孟亚轩, 赵心月, 等.谷子bHLH转录因子家族基因鉴定及生物信息学分析[J].种子, 2021, 40(12):45-55.
SUN Y Q, MENG Y X, ZHAO X Y, et al.Identification and bioinformatics analysis of transcription factor family genes bHLH in foxtail millet[J].Seed, 2021, 40(12):45-55.
[31] FAN W G, LI B Y, TIAN H, et al.Metabolome and transcriptome analysis predicts metabolism of violet-red color change in Lilium bulbs[J].Journal of the Science of Food and Agriculture, 2022, 102(7):2903-2915.