单端孢霉烯族毒素(trichothecenes,TCT)是由镰刀菌属(Fusarium)等真菌产生的一类化学结构相似的次级代谢产物的统称[1]。镰刀菌广泛分布于土壤和有机体中,具有较强的环境适应性、抗逆性和侵染能力,生长过程中极易产生真菌毒素且产毒种类丰富,毒性较强[2]。我国地域广阔,气候多样,大部分粮食产区属于温带季风气候,气候温和,降雨丰沛,为某些真菌的生长提供了理想的条件,可能导致农作物的真菌毒素污染问题,长期摄入含有TCT的食物不仅严重威胁人畜健康,还给粮食、饲料生产、畜禽养殖等造成巨大经济损失[3]。本文简要介绍TCT的污染现状和危害,系统总结近年来TCT的生物降解新进展,并对其降解机理展开讨论,为安全高效地去除TCT提供理论参考。
1 单端孢霉烯族毒素的基本结构和分类
TCT指主要由镰刀菌、头孢霉(Cephalosporium)、漆斑菌(Myrothecium)、葡萄穗霉(Stachybotrys)、木霉(Trichoderma)等真菌产生的一类有毒四环倍半萜化合物,已发现200多种,根据结构差异可分为A型、B型、C型、D型四类[4]。其中,A型和B型是食品和饲料污染情况最严重、毒性作用最强的2种类型[5],它们以C-8位是否含有羰基加以区分(表1)。其中,A型以T-2毒素(T-2 toxin,T-2)、HT-2毒素(HT-2 toxin,HT-2)、新茄镰孢菌醇(neosolaniol,NEO)、蛇形菌素(diacetoxyscirpenol,DAS)为代表;B型则以脱氧雪腐镰刀菌烯醇(deoxynivalenol,DON)、雪腐镰刀菌烯醇(nivalenol,NIV)和镰刀菌烯酮(fusarenon-X,FUS)为代表,其中还包括3-乙酰脱氧雪腐镰刀菌烯醇(3-acetyldeoxynivalenol,3-AcDON)和15-乙酰脱氧雪腐镰刀菌烯醇(15-acetyldeoxynivalenol,15-AcDON)这2种DON的衍生物[5]。
表1 A、B型单端孢霉烯族毒素及其结构
Table 1 Type A and B trichothecenes and their respective structures
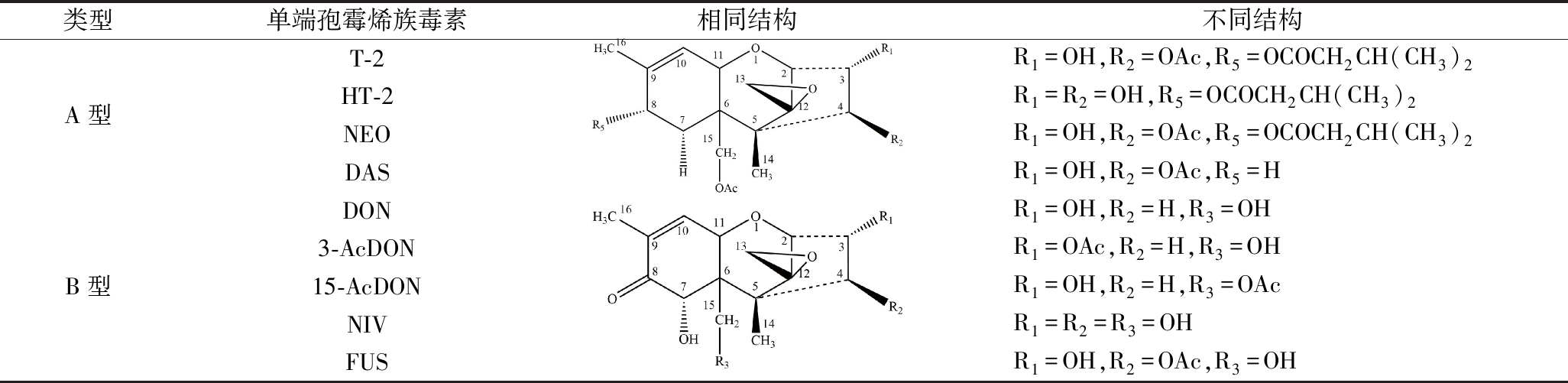
类型单端孢霉烯族毒素相同结构不同结构A型T-2HT-2NEODASR1=OH,R2=OAc,R5=OCOCH2CH(CH3)2R1=R2=OH,R5=OCOCH2CH(CH3)2R1=OH,R2=OAc,R5=OCOCH2CH(CH3)2R1=OH,R2=OAc,R5=HB型DON3-AcDON15-AcDONNIVFUSR1=OH,R2=H,R3=OHR1=OAc,R2=H,R3=OHR1=OH,R2=H,R3=OAcR1=R2=R3=OHR1=OH,R2=OAc,R3=OH
2 单端孢霉烯族毒素的污染现状及危害
2.1 单端孢霉烯族毒素的污染现状
TCT的污染主要发生于作物的生长期和贮藏期,对其污染情况调查发现,从美国14个州收集的707个猪饲料样品中DON、3-AcDON、15-AcDON和NIV的检出率分别为94%、33%、57%和1%[6]。KOSICKI等[7]检测了从波兰收集的60个冬黑麦样品中镰刀菌毒素(DON、NIV、3-AcDON、DAS、T-2、HT-2等)的污染情况,90%的样品存在DON,T-2毒素和HT-2毒素的检出率分别为63%和57%。从阿根廷北部采集的新收获小麦样本中发现3-AcDON、HT-2和DON的检出率分别为21.7%、22.5%和85%[8]。乌拉圭西南部的4个小麦产区采集的小麦样本中70%的样本DON呈阳性[9]。真菌的生长和毒素合成受温度、湿度等环境因素的影响[10],温暖潮湿的环境更利于产生TCT。研究表明燕麦开花期间和开花前后温暖、多雨和潮湿的天气增加了DON积累的风险,幼苗生长和分蘖期也会出现相同现象[11]。此外,TCT的污染程度也根据作物种类的不同而存在明显差异,比如小麦和玉米等易受赤霉病等侵袭,它们受到TCT污染的风险远高于其他作物。对2017—2021年我国饲料样品中真菌毒素调查发现,相较于豆粕中的TCT阳性检出率为14.79%,玉米中B型TCT的阳性检出率则高达84.04%[12]。研究发现英国、法国和埃及等地区孕妇尿样中存在含量较高的DON及其代谢物[13],这表明DON可通过食物链进入人体,并对孕妇及其胎儿的健康产生影响。
2.2 单端孢霉烯族毒素的危害
TCT不仅可以抑制蛋白质和核酸合成,破坏线粒体功能、造血系统和免疫系统,对人和动物造成急慢性疾病,如呕吐、腹泻、器官出血性坏死、胃肠道炎症以及免疫失调等[14],还能抑制生长激素的合成和转运,从而影响动物生长发育[15]。TCT主要通过粮食、饲料及其加工制品的直接或间接摄入,在人和动物体内蓄积、代谢和转化,损伤消化道黏膜,引起胃肠炎症,降低免疫机能。胃肠道是抵御食源性污染物的第一道生理屏障,TCT被摄入后,可通过改变胃肠道细胞形态和分化以及屏障功能来影响胃肠功能[16]。T-2毒素可诱导与炎症反应相关的肠功能损伤,并损害猪肠上皮细胞的肠屏障功能[17],高剂量摄入会引起免疫器官和造血细胞坏死、造成生殖和胚胎毒性[18-20]。B型TCT是TCT中污染最广泛、毒性最强的一类,其中又以DON最为严重,动物摄入含DON的食物会造成恶心、呕吐、食欲衰退、贫血、免疫力下降和内分泌失调等症状,具有致癌、致畸及免疫毒性等[21]。
2.3 单端孢霉烯族毒素的限量标准
随着研究的不断深入,世界各国和地区相继制定、修订了相应限量标准,目前,TCT的限量主要集中于T-2毒素和DON。在我国,GB 2761—2017《食品中真菌毒素限量》中规定谷物及其制品中DON的限量为1 000 μg/kg;GB 13078—2017《饲料卫生标准》中规定植物性饲料原料中DON限量为5 000 μg/kg,饲料原料中T-2毒素的限量为500 μg/kg;美国食品药品监督管理局(Food and Drug Administration,FDA)规定食品中的DON的安全标准是1 000 μg/kg;国际食品法典委员会(Codex Alimentarius Commission,CAC)CODEX STAN 193—2010中对小麦、玉米和大麦中DON的限量为2 000 μg/kg,对三者制成的面粉、粗粉或麦片限量为1 000 μg/kg;欧盟(European Union,EU)(EC) No.1881/2006中规定未加工的小麦、玉米和燕麦限量为1 250 μg/kg,供消费者食用的谷物、谷物粉、粗面粉、麸皮和麦芽中DON限量为750 μg/kg,较CAC和我国更为严格。
3 单端孢霉烯族毒素的生物降解
生物降解是在温和条件下利用微生物或生物酶将真菌毒素转化为无毒或低毒产物,不会造成粮食和饲料的营养损失和二次污染,具有绿色安全等优点。目前,TCT生物降解的研究涵盖微生物单独降解、复合菌群协同降解及生物酶降解。
3.1 细菌降解
通过富集纯化等方法筛选出具有降解TCT效果的细菌,可以有效地降低TCT的毒性。目前针对TCT降解细菌的研究主要集中于DON和T-2毒素两类。
一些从特定环境分离的细菌可以将DON降解为毒性较低的代谢产物。FUCHS等[22]从牛瘤胃中分离的真杆菌属(Eubacterium sp.)BBSH797菌株可将DON脱环氧化,而BBSH797需要严格厌氧培养,这无疑限制了该菌的应用范围;HE等[23]从土壤中筛选的德沃斯氏菌(Devosia sp.)17-2-E-8对DON有较高的降解效果3 μg DON/(h/108个细胞),并成功分离出关键降解酶,为后续酶制剂制备开发提供可能;WANG等[24]分离出的Ketogulonicigenium vulgare D3_3在有氧或无氧条件下均能有效降解DON;IKUNAGA等[25]和ZHANG等[26]从土壤中分离的类诺卡氏菌(Nocardioides sp.)可将DON转化生成3-epi-DON,该菌还具有异养同化DON的能力;此外,一些芽孢杆菌也具有DON降解功能,芽孢杆菌(Bacillus sp.)LS100不仅可有效去除谷物中污染DON,还不会改变谷物的营养和风味[27]。
除了发现DON降解细菌,还有许多关于T-2毒素降解细菌的报道。蜡样芽胞杆菌(Bacillus cereus)[28]和暹逻芽孢杆菌(Bacillus siamensis)B26[29]对T-2毒素降解效果均超过60%。除了芽孢杆菌外,解脂厌氧弧菌(Anaerovibrio lipolytica)、反刍月形单胞菌(Selenomonas ruminantium)等均可降解T-2毒素[30]。这些细菌的发现对探索TCT的降解新途径提供生物资源。
3.2 真菌降解
目前报道的能降解TCT的真菌主要有酵母(yeast)、曲霉(Aspergillus)、木霉(Trichoderma)和青霉(Penicillium)等。一些酵母不仅在食品和饲料的发酵中发挥重要作用,还具有降解DON和T-2毒素的能力,邵春山等[31]从陈年普洱茶叶中分离的毕赤酵母(Pichia sp.)MC-1可降解豆粕中46.79%的DON;MCCORMICK等[32]发现的酵母菌可将T-2毒素进行不同的生物转化:乙酰化为3-乙酰基T-2毒素、糖基化为T-2毒素3-葡萄糖苷或去除异戊基形成NEO。目前发现的木霉和青霉主要对DON有降解效果,TIAN等[33]发现木霉(Trichoderma harzianum)Q710613,(Trichoderma atroviride)Q710251和(Trichoderma asperellum)Q710682不仅可以抑制禾谷镰刀菌(Fusarium graminearum)生长,减少真菌毒素产生,还可以将DON转化成脱氧雪腐镰刀菌烯醇-3-葡萄糖苷(DON-3-glucoside,D3G);李亚菲等[34]筛选出的青霉(Penicillium sp.)Ma-1-4可降解玉米秸秆中46%的DON;曲霉作为发酵工业和食品加工业的重要菌种,部分菌株具有DON和T-2毒素降解功能,如米曲霉(Aspergillus oryzae)培养21 d可以降解92%的DON[35],而黑曲霉(Aspergillus Niger)菌株在14 d可以降解90%的T-2毒素[36]。
3.3 混合菌群降解
由于单一菌株对不同毒素的敏感程度不同或者需要特定菌株产生的辅酶来共同辅助降解,因此对于复杂基质中的毒素,单一菌株降解效果不理想时可以采用混合菌种复配来实现更好的效果。将枯草芽孢杆菌、酿酒酵母菌、丁酸梭菌3种益生菌进行有效配伍,不仅可以提高DON的降解效果,而且能同步降解黄曲霉毒素B1和玉米赤霉烯酮[37];WANG等[38]筛选出由假单胞菌和德沃斯氏菌组成的混合菌群能完全降解DON,其中假单胞菌提供辅酶吡咯喹啉醌(pyrroloquinoline quinone disodium salt,PQQ)协同德沃斯氏菌中脱氢酶降解DON;另外,经过选择性培养和驯化筛选出的微生物群落可用于降解多种毒素,例如WANG等[39]从土壤中筛选出的细菌群落C20可降解15-AcDON、3-AcDON和T-2毒素;HE等[40]采用原位富集法从土壤中分离出的细菌群落PGC-3可将DON和NIV完全脱环氧化。不论是多种降解菌株协同作用还是为降解菌株提供必要的辅酶,混合菌种在降解毒素方面都有一定的研究价值,但在降解过程中如何保证复合菌群的稳定性等问题还有待深入研究。
3.4 生物酶降解
相较于降解菌株筛选,针对TCT降解酶的研究进展目前还极为有限,而且多集中于DON和T-2毒素中。CARERE等[41]对Devosia spp. 17-2-E-8中DON降解酶进行挖掘,突破性发现PQQ依赖的脱氧雪腐镰刀菌烯醇差向异构化酶(A)(deoxynivalenol epimerization enzyme(A),DepA)和还原型烟酰胺腺嘌呤二核苷酸磷酸(reduced nicotinamide adenine dinucleotide phosphate,NADPH)依赖的脱氢酶DepB[42]可将DON先氧化为3-keto-DON再还原为DON的同分异构体3-epi-DON。HE等[43]从鞘氨醇单胞菌(Sphingomonas sp.)S3-4中克隆出醛酮还原酶AKR18A1,该酶在辅酶氧化型烟酰胺腺嘌呤二核苷酸磷酸(Electron acceptors nicotinamide adenine dinucleotide phosphate,NADP+)催化下可将DON氧化为3-oxo-DON。研究学者通过计算机辅助筛选,结合生物信息学预测以及酶的体外表达等手段相继发现与DepA功能相似的脱氧雪腐镰刀菌烯醇脱氢酶(deoxynivalenol dehydrogenase,DDH)[44]、醌依赖性脱氧雪腐镰刀菌烯醇脱氢酶(quinone-dependent deoxynivalenol dehydrogenase,QDDH)[45]。、山梨糖脱氢酶(sorbitol dehydrogenase,SDH)[46]和YoDDH[47]等,以及与DepB功能相似醛酮还原酶AKR13B2和AKR6D1[44]等;部分研究学者对已发现的降解酶进行优化改造提高其降解能力,如定点突变DDH得到突变脱氧雪腐镰刀菌烯醇脱氢酶(mutational deoxynivalenol dehydrogenase,TDDH)可提高酶的降解性能[44],SDH的突变体F103L和F103A可通过扩大活性口袋提高对DON的降解活性[46]。
除了微生物来源的酶,研究人员还对动物体内的酶系开展了相关研究,发现羧酸酯酶和细胞色素酶家族等对TCT也有一定降解效果,如人肝微粒体中羧酸酯酶[48]和白细胞中羧酸酯酶[49]可以降解T-2毒素。与羧酸酯酶类似,动物肝细胞中发现的细胞色素P450酶系统也可以降解T-2毒素[50]。
植物体内也发现可降解DON和T-2毒素的生物酶。其中,漆酶被认为是“绿色催化剂”,可以通过漆酶介导体系来氧化TCT,天然介体乙酰丁香酮(acetosyringone,AS)、丁香醛(syringaldehyde,SA)与漆酶(Lac-W)构建的Lac-W-AS体系和Lac-W-SA体系可以降解DON,降解率分别为43%和28%[51]。植物体产生的过氧化物酶通过强大高效的氧化能力可将毒素基本骨架完全破坏,如从蘑菇中提取的木质素过氧化物酶(Lignin peroxidase,LiP)和锰过氧化物酶(Mn-dependent peroxidase,MnP)均可降解DON[52];从米糠中纯化的过氧化物酶可使DON减少81.7%[53]。
目前,多数筛选到的TCT降解菌株的降解机制尚不明确,针对新型TCT类真菌毒素降解酶的挖掘手段较为有限,因此,亟需针对新型降解酶的挖掘开展深入系统的研究,为真菌毒素脱毒剂的开发和脱毒技术的推广应用提供支撑。
4 单端孢霉烯族毒素的生物降解机理
目前已经针对TCT开展了一些降解研究,但多数降解机理不清,降解产物不明,严重制约了相关脱毒产品的开发和脱毒技术的产业化应用。在此,本文系统地梳理了TCT生物转化相关的研究进展,以期为TCT生物降解新机制的发现和新产物的结构解析提供借鉴。
4.1 羟基化和羰基化
生物转化去除或转移TCT的羟基来降低其毒性的脱毒方式在DON和T-2毒素方面研究较多。研究发现德沃斯氏菌与PQQ共同作用能将DON C3位上的羟基氧化成3-keto-DON[41],再与NADPH反应将3-keto-DON还原为3-epi-DON[42];LI等[46]通过分子对接和稳定性预测筛选出的SDH可以将DON降解为3-keto-DON,同时推测另一个产物可能将DON的2个羟基转化为酮基;SHI等[47]利用Enzyme Miner筛选出新的DON降解酶YoDDH可同时利用电子受体PQQ、吩嗪硫酸甲酯和2, 6-二氯苯酚吲哚酚来氧化DON生成3-keto-DON;研究发现生物体中细胞色素P450可以参与外源性物质如TCT的代谢,ITO等[54]从鞘氨醇单胞菌Sphingomonas sp.KSM1的基因组文库中筛选出编码细胞色素P450的基因ddnA,并从KSM1亲缘较近的菌株中克隆出核黄素腺嘌呤二核苷酸(flavin adenine dinucleotide,FAD)依赖性铁氧还蛋白还原酶Kdx和线粒体型[2Fe-2S]铁氧还蛋白KdR的编码基因,得到重组蛋白DdnA-Kdx-KdR与还原型辅酶Ⅰ(nicotinamide adenine dinucleotide,NADH)组成的Ⅰ类P450系统可将DON的C16甲基羟基化生成16-羟基-DON(16-hydroxy-deoxynivalenol,16-HDON),该系统对NIV和3-AcDON也有降解活性(图1-a)。KOBAYASHI等[55]研究发现细胞色素P450可以将T-2毒素和HT-2毒素羟基化,生成3′-羟基-T-2毒素(3′-OH-T-2)和3′-羟基HT-2毒素(3′-OH-HT-2)(图1-b)。
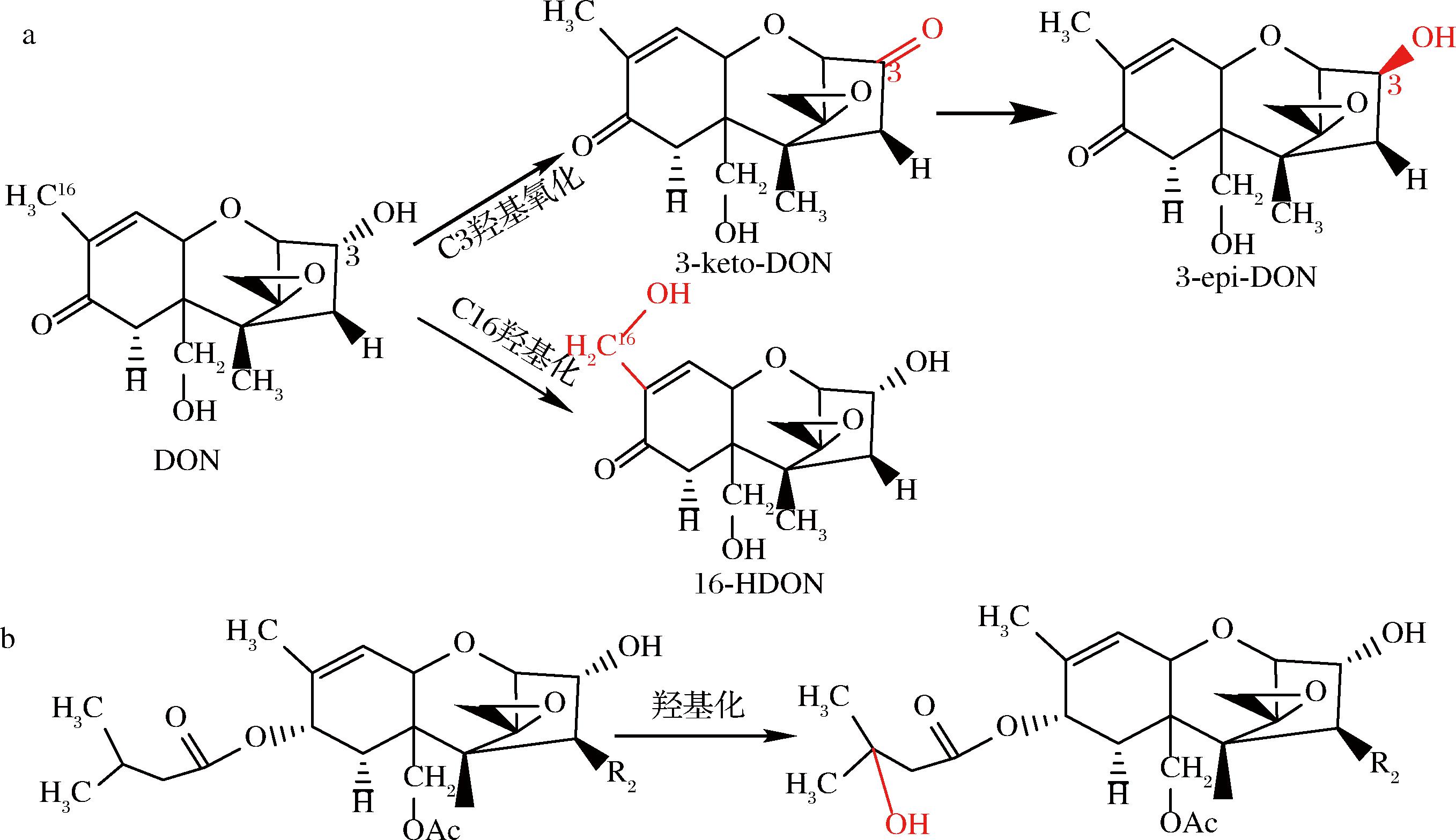
a-DON的羟基化反应;b-T-2(R2=OAc)和HT-2(R2=OH)羟基化反应
图1 DON、T-2和HT-2羟基化反应
Fig.1 DON、T-2 and HT-2 hydroxylation reactions
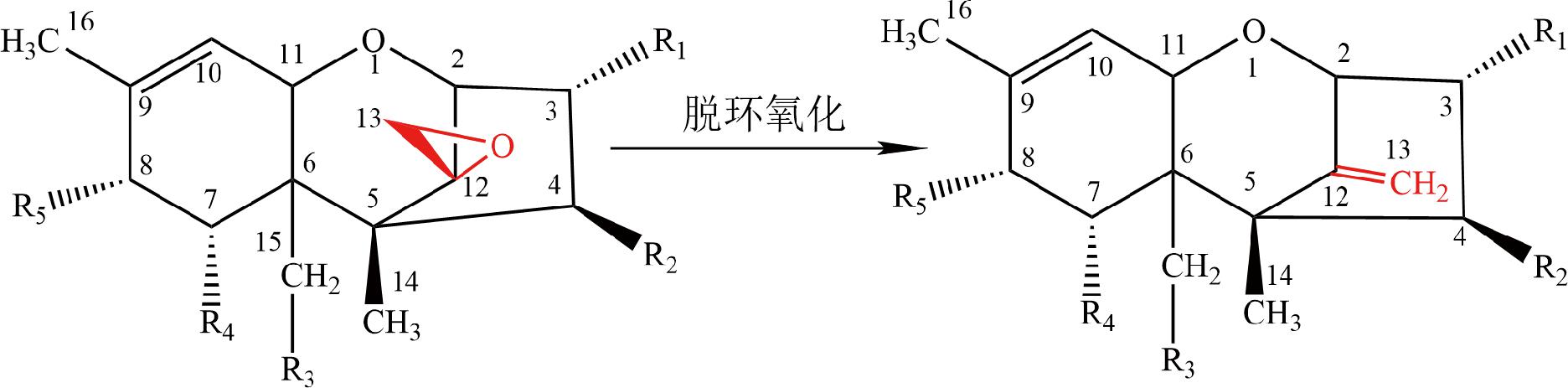
图2 脱环氧化反应
Fig.2 Deepoxidation reaction
4.2 脱环氧化
微生物可以破坏TCT的C12、C13环氧结构降低其毒性。SWANSON等[56]研究发现从牛和猪等动物体内分离的微生物可以使DAS完全脱环氧化生成脱环氧单乙酰镰刀菌烯三醇(deepoxy monoacetoxyscirpenol,DE MAS)和脱环氧镰刀菌烯三醇(deepoxy scirpentriol,DE SCP)。作为商业化饲料添加剂的菌株BBSH797可将DON、HT-2毒素和T-2三醇脱环氧化[22],从鸡肠道分离出的伊格尔兹氏菌(Eggerthella sp.)DII-9[57]、梭菌属(Clostridium sp.)WJ06[58]和史雷克氏菌(Slackia sp.)D-G6[59]也具有相似的脱环氧化功能。动物胃肠道是严格的厌氧条件,降解菌株对TCT脱环氧化的过程需要严格厌氧,而这给实际应用增加了难度。同时,有研究发现有些菌株可在有氧条件下将TCT脱环氧化,HE等[60]分离的脱硫杆菌(Desulfitobacterium) sp.PGC-3-9能够在有氧条件下将HT-2、DON、NIV和15-AcDON脱环氧化,但厌氧条件下效果更好。
4.3 (水解)脱乙酰基化
C12,13环氧环和C3羟基是TCT毒性作用的主要基团,但乙酰基的位置和数量对TCT的毒性和生物转化也有一定程度的影响。YOUNG等[61]从鸡肠道消化物中分离的混合微生物或分离物LS100和SS3可将3-AcDON、15-AcDON和FUS脱乙酰基化。UENO[62]和马妍等[63]分别发现短小杆菌114-2和菌株AFJ-2可将T-2毒素水解为HT-2毒素后再脱去C15位乙酰基转化成T-2三醇。马妍等[63]还发现菌株AFJ-2和AFJ-3共同作用于T-2毒素可产生新的产物4-脱乙酰-NEO。溶纤维丁酸弧菌(Butyrivibrio fibrisolvens) M-4a可将DAS脱乙酰基形成15-乙酰镰刀菌烯三醇(15-acetoxyscirpenol,15-ASCP)[64]。研究发现动物的组织与细胞中存在胃蛋白酶、胰酶等一系列可以脱乙酰基的活性物质,对3-AcDON和15-AcDON有较强的脱乙酰能力[65]。
4.4 糖基化作用
生物体为降低TCT产生的毒性通过自身防御机制与葡萄糖醛酸苷、葡萄糖苷等物质发生共轭反应形成糖基化产物。DON糖基化反应易发生于C3、C15位,生成DON-3-葡萄糖醛酸苷(DON-3-glucoside,DON-3-O-GlcAc)和DON-15-葡萄糖醛酸苷(DON-15-glucoside,DON-15-O-GlcAc)[66];从水稻中克隆的糖基转移酶能够转化DON生成D3G[67],(图3-a)。拟南芥中DON-糖基转移酶Ⅰ可以催化葡萄糖从二磷酸尿苷-葡萄糖(uridine diphosphate,UDP)转移到DON的C3羟基,形成3-β-D-吡喃葡萄糖基-4-DON,这种酶的过表达加强了拟南芥对DON的抵抗能力[68];而动物体中的尿苷二磷酸-葡萄糖基转移酶(uridine diphosphate glucuronosyltransferase,UGT)也有相似功能,如大鼠肝微粒体中UGT能将DON糖苷酸化[69],从而降低DON对大鼠肝脏器官的危害;WELSCH等[70]优化肝微粒体UGT合成T-2和HT-2毒素葡萄糖醛酸苷的方法,并鉴定出HT-2毒素3-葡萄糖醛酸苷(HT-2 toxin 3-glucuronide)的新异构体HT-2毒素4-葡萄糖醛酸苷(HT-2 toxin 4-glucuronide)(图3-b)。
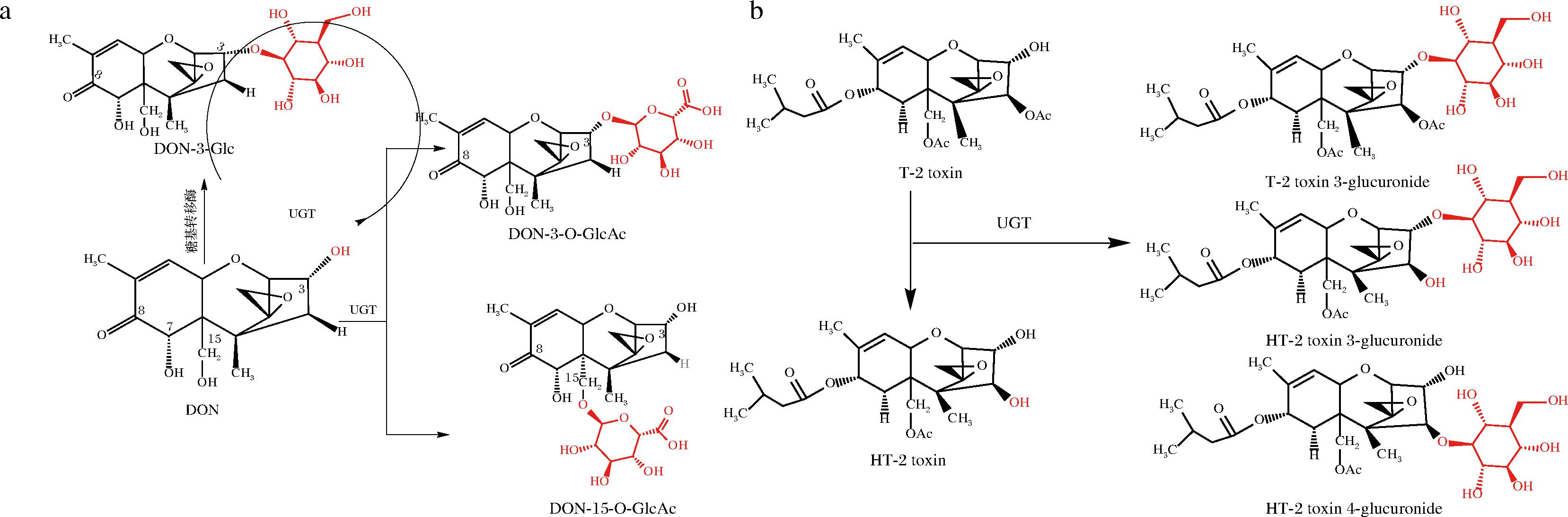
a-DON糖基化反应;b-T-2、HT-2毒素糖基化反应
图3 DON、T-2和HT-2糖基化反应[70]
Fig.3 DON,T-2 and HT-2 glycosylation reactions[70]
4.5 其他作用
随着技术发展,越来越多TCT的降解机制被不断发现,但仍有许多降解途径和影响因素尚不清晰,降解产物结构和安全性不明。因此,对降解机制的解析至关重要。对于已发现的一些降解菌株,研究者也结合实验结果解析其降解机制,如曲霉属菌株(Aspergillus tubingensis) NJA-1利用环氧化物水解酶(epoxide hydrolase,EH)可水解DON的环氧环[71]。研究人员还发现真菌可以通过将毒素转化为低毒物质从而实现自我保护,如禾谷镰刀菌通过乙酰化T-2毒素,HT-2毒素和NEO的羟基来降低毒素毒性[72];真菌漆酶在2,2,6,6-四甲基哌啶-N-氧基(2,2,6,6-tetramethylpiperidinooxy, TEMPO)存在时,将DON的C3和C15位羟基氧化成酮,然后TEMPO连接到C4位形成3,15-diketo,4-TEMPO-DON[73]。目前报道的降解菌中,还有类诺卡氏菌(Nocardioides sp.)WSN05-2[25]和ZHH-013[26]能将DON转化,但相应的转化机制并未完全解析。此外,木质素过氧化物酶和锰过氧化物酶也展现出较好能DON降解效果[52],但降解产物和降解机制尚不清楚,可能是其强氧化力导致毒素基本骨架过度破坏或者与介体相互作用形成未知的复杂化合物。
5 展望
近年来,针对真菌毒素生物降解的研究取得了很多可喜的进展,但是利用生物降解来减少或消除食品和饲料中TCT污染的研究还十分有限,可以针对以下几个方面开展系统、深入的研究:a)目前对TCT降解机制的研究相对不足,深入探究降解机理,明确反应条件以及降解产物的毒理学评价,有助于确定关键的降解限速步骤和限制因素,从而通过条件优化等方式提高降解效率和应用范围;b)利用新型生物酶挖掘技术发掘TCT降解酶,并结合人工智能高效推进降解酶结构修饰和分子改造,促进降解酶改良提效,实现降解酶制剂的工业化应用;c)当前部分降解过程需辅酶协助催化,辅酶价格昂贵,使用成本较高。可以根据降解机理,进行辅酶再生或替代,以实现辅酶自给自足,从而提高生物降解技术的经济可行性和实用性。
[1] DESJARDINS A E, HOHN T M, MCCORMICK S P.Trichothecene biosynthesis in Fusarium species:Chemistry, genetics, and significance[J].Microbiological Reviews, 1993, 57(3):595-604.
[2] 马忠华, 陈云, 尹燕妮.小麦赤霉病流行成灾原因分析及防控对策探讨[J].中国科学基金, 2020, 34(4):464-469.
MA Z H, CHEN Y, YIN Y N.Epidemiological analysis and management strategies of Fusarium head blight of wheat[J].Bulletin of National Natural Science Foundation of China, 2020, 34(4):464-469.
[3] 王刚, 王玉龙, 张海永, 等.真菌毒素形成的影响因素[J].菌物学报, 2020, 39(3):477-491.
WANG G,WANG Y L,ZHANG,H Y, et al.Factors that affect the formation of mycotoxins:A literature review[J].Mycosystema, 2020, 39(3):477-491.
[4] MCCORMICK S P, STANLEY A M, STOVER N A, et al.Trichothecenes:From simple to complex mycotoxins[J].Toxins, 2011, 3(7):802-814.
[5] MACRI A M, NAGY A L, DAINA S, et al.Occurrence of types A and B trichothecenes in cereal products sold in Romanian markets[J].Toxins, 2023, 15(7):466.
[6] PACK E D, WEILAND S, MUSSER R, et al.Schmale DG.Survey of zearalenone and type-B trichothecene mycotoxins in swine feed in the USA[J].Mycotoxin Research.2021, 37(4):297-313.
[7] KOSICKI R, ![]() M, DOPIERA
M, DOPIERA A P, et al.Occurrence of mycotoxins in winter rye varieties cultivated in Poland (2017-2019)[J].Toxins, 2020, 12(6):423.
A P, et al.Occurrence of mycotoxins in winter rye varieties cultivated in Poland (2017-2019)[J].Toxins, 2020, 12(6):423.
[8] GONZ LEZ H L, MOLT
LEZ H L, MOLT G A, PACIN A, et al.Trichothecenes and mycoflora in wheat harvested in nine locations in Buenos Aires province, Argentina[J].Mycopathologia, 2008, 165(2):105-114.
G A, PACIN A, et al.Trichothecenes and mycoflora in wheat harvested in nine locations in Buenos Aires province, Argentina[J].Mycopathologia, 2008, 165(2):105-114.
[9] PAN D, GRANERI J, BETTUCCI L.Correlation of rainfall and levels of deoxynivalenol in wheat from Uruguay, 1997-2003[J].Food Additives &Contaminants.Part B, Surveillance, 2009, 2(2):162-165.
[10] MÜLLER M E H, BRENNING A, VERCH G, et al.Multifactorial spatial analysis of mycotoxin contamination of winter wheat at the field and landscape scale[J].Agriculture, Ecosystems &Environment, 2010, 139(1-2):245-254.
[11] HJELKREM A R, TORP T, BRODAL G, et al.DON content in oat grains in Norway related to weather conditions at different growth stages[J].European Journal of Plant Pathology, 2017, 148(3):577-594.
[12] HAO W, GUAN S, LI A P, et al.Mycotoxin occurrence in feeds and raw materials in China:A five-year investigation[J].Toxins, 2023, 15(1):63.
[13] MISHRA S, SRIVASTAVA S, DEWANGAN J, et al.Global occurrence of deoxynivalenol in food commodities and exposure risk assessment in humans in the last decade:A survey[J].Critical Reviews in Food Science and Nutrition, 2020, 60(8):1346-1374.
[14] ROCHA O, ANSARI K, DOOHAN F M.Effects of trichothecene mycotoxins on eukaryotic cells:A review[J].Food Additives and Contaminants, 2005, 22(4):369-378.
[15] MAHATO D K, PANDHI S, KAMLE M, et al.Trichothecenes in food and feed:Occurrence, impact on human health and their detection and management strategies[J].Toxicon, 2022, 208:62-77.
[16] PINTON P, OSWALD I P.Effect of deoxynivalenol and other type B trichothecenes on the intestine:A review[J].Toxins, 2014, 6(5):1615-1643.
[17] HE W H, WANG J H, HAN M Y, et al.Potential toxicity and mechanisms of T-2 and HT-2 individually or in combination on the intestinal barrier function of porcine small intestinal epithelial cells[J].Toxins, 2023, 15(12):682.
[18] 吴朝金, 王雅玲, 孙力军, 等.对虾中T-2毒素对小鼠免疫功能和血清生化指标的影响[J].中国食品学报, 2015, 15(5):166-174.
WU C J, WANG Y L, SUN L J, et al. Effect of T-2 toxin in litopenaeus vannamei on immune and serum biochemical indexe of mice[J]. Journal of Chinese Institute of Food Science and Technology, 2015, 15(5):166-174.
[19] DOI K,ISHIGAMI N,SEHATA S.T-2 toxin-induced toxicity in pregnant mice and rats[J].International Journal of Molecular Sciences,2008,9(11):2146-2158.
[20] FU X Y,GAO Y H, YANG Y M, et al.Induced lesion and inhibited Ihh-PTHrP signalling pathway activity in the articular cartilage of rats caused by T-2 toxin[J].Toxicon,2019,158:104-108.
[21] GAROFALO M, PAYROS D, PENARY M, et al.A novel toxic effect of foodborne trichothecenes:The exacerbation of genotoxicity[J].Environmental Pollution, 2023, 317:120625.
[22] FUCHS E, BINDER E M, HEIDLER D, et al.Structural characterization of metabolites after the microbial degradation of type A trichothecenes by the bacterial strain BBSH 797[J].Food Additives and Contaminants, 2002, 19(4):379-386.
[23] HE J W, HASSAN Y I, PERILLA N,et al.Bacterial epimerization as a route for deoxynivalenol detoxification:The influence of growth and environmental conditions[J].Frontiers in Microbiology, 2016, 7:572.
[24] WANG Y, ZHAO D L, ZHANG W, et al.Four PQQ-dependent alcohol dehydrogenases responsible for the oxidative detoxification of deoxynivalenol in a novel bacterium Ketogulonicigenium vulgare D3_3 originated from the feces of Tenebrio molitor larvae[J].Toxins, 2023, 15(6):367.
[25] IKUNAGA Y, SATO I, GROND S, et al.Nocardioides sp.strain WSN05-2, isolated from a wheat field, degrades deoxynivalenol, producing the novel intermediate 3-epi-deoxynivalenol[J].Applied Microbiology and Biotechnology, 2011, 89(2):419-427.
[26] ZHANG H H, ZHANG H, QIN X, et al.Biodegradation of deoxynivalenol by Nocardioides sp.ZHH-013:3-keto-deoxynivalenol and 3-epi-deoxynivalenol as intermediate products[J].Frontiers in Microbiology, 2021, 12:658421.
[27] LI X Z, ZHU C, DE LANGE C M, et al.Efficacy of detoxification of deoxynivalenol-contaminated corn by Bacillus sp.LS100 in reducing the adverse effects of the mycotoxin on swine growth performance[J].Food Additives &Contaminants.Part A, Chemistry, Analysis, Control, Exposure &Risk Assessment, 2011, 28(7):894-901.
[28] 周浪花, 王雅玲, 张春辉, 等.对虾肠道中T-2毒素降解菌的分离纯化与鉴定[J].微生物学杂志, 2017, 37(6):50-56.
ZHOU L H,WANG Y L,ZHANG C H,et al.Isolation and identification of T-2 toxin-degradable strains from shrimp intestine[J].Journal of Microbiology, 2017, 37(6):50-56.
[29] 向雨珂, 熊犍, 张晓琳, 等.T-2毒素脱毒菌株的筛选及脱毒机制初探[J].食品科技, 2017, 42(11):27-33.
XIANG Y K, XIONG J, ZHANG X L, et al.Screening of T-2 toxin detoxification strains and the mechanism of detoxification[J].Food Science and Technology,2017, 42(11):27-33.
[30] WESTLAKE K, MACKIE R I, DUTTON M F.T-2 toxin metabolism by ruminal bacteria and its effect on their growth[J].Applied and Environmental Microbiology, 1987, 53(3):587-592.
[31] 邵春山, 余祖华, 廖成水, 等.一株毕赤酵母菌 MC-1 降解呕吐毒素、生物学特性及初步应用[J].中国饲料, 2023(3):37-43.
SHAO C S, YU Z H, LIAO C S, et al. A strain of Pichia pastoris Mc-1 degrades vomiting toxin, its biological characteristics and its preliminary application[J]. China Feed, 2023(3):37-43.
[32] MCCORMICK S P, PRICE N P J, KURTZMAN C P.Glucosylation and other biotransformations of T-2 toxin by yeasts of the Trichomonascus clade[J].Applied and Environmental Microbiology, 2012, 78(24):8694-8702.
[33] TIAN Y, TAN Y L, LIU N, et al.Detoxification of deoxynivalenol via glycosylation represents novel insights on antagonistic activities of Trichoderma when confronted with Fusarium graminearum[J].Toxins, 2016, 8(11):335.
[34] 李亚菲, 余祖华, 刘赛宝, 等.一株降解呕吐毒素的青霉菌的分离与鉴定[J].饲料工业, 2015, 36(15):42-45.
LI Y F,YU Z H,LIU S B,et al.Isolation and identification of a Penicillium strain degraded deoxynivalenol[J].Feed Industry,2015, 36(15):42-45.
[35] TRAN S T, SMITH T K.Conjugation of deoxynivalenol by Alternaria alternata (54028 NRRL), Rhizopus microsporus var.rhizopodiformis (54029 NRRL) and Aspergillus oryzae (5509 NRRL)[J].Mycotoxin Research 2014, 30(1):47-53.
[36] 吴娱. T-2毒素降解菌的筛选、降解酶的提取及其降解效果的研究[D].合肥:合肥工业大学, 2016.
WU Y.Screening of T-2 toxin degrading bacteria, extraction of degrading enzyme and its degradation effect[D].Hefei:Hefei University of Technology, 2016.
[37] 王晓敏, 常娟, 王平, 等.复合益生菌和霉菌毒素降解酶对黄曲霉毒素B1、玉米赤霉烯酮和呕吐毒素的同步降解[J].中国饲料, 2021(21):85-91.
WANG X M, CHANG J, WANG P, et al.Synchronous degradation of aflatoxin B1, Zearalenone and donxynivalenol by compound probiotics and mycotoxin-degradation enzymes[J].China Feed, 2021(21):85-91.
[38] WANG Y, ZHAO D L, ZHANG W, et al.Biotransformation of deoxynivalenol by a dual-member bacterial consortium isolated from Tenebrio molitor larval feces[J].Toxins, 2023, 15(8):492.
[39] WANG Y X, WANG G, DAI Y J, et al.Biodegradation of deoxynivalenol by a novel microbial consortium[J].Frontiers in Microbiology, 2020, 10:2964.
[40] HE W J, YUAN Q S, ZHANG Y B, et al.Aerobic de-epoxydation of trichothecene mycotoxins by a soil bacterial consortium isolated using in situ soil enrichment[J].Toxins, 2016, 8(10):277.
[41] CARERE J, HASSAN Y I, LEPP D, et al.The enzymatic detoxification of the mycotoxin deoxynivalenol:Identification of DepA from the DON epimerization pathway[J].Microbial Biotechnology, 2018, 11(6):1106-1111.
[42] CARERE J, HASSAN Y I, LEPP D, et al.The identification of DepB:An enzyme responsible for the final detoxification step in the deoxynivalenol epimerization pathway in Devosia mutans 17-2-E-8[J].Frontiers in Microbiology, 2018, 9:1573.
[43] HE W J, ZHANG L M, YI S Y, et al.An aldo-keto reductase is responsible for Fusarium toxin-degrading activity in a soil Sphingomonas strain [J].Scientific Reports, 2017, 7(1):9549.
[44] QIN X J, ZHANG J, LIU Y R, et al.A quinoprotein dehydrogenase from Pelagibacterium halotolerans ANSP101 oxidizes deoxynivalenol to 3-keto-deoxynivalenol[J].Food Control, 2022, 136:108834.
[45] HE W J, SHI M M, YANG P, et al.A quinone-dependent dehydrogenase and two NADPH-dependent aldo/keto reductases detoxify deoxynivalenol in wheat via epimerization in a Devosia strain[J].Food Chemistry, 2020, 321:126703.
[46] LI D Y, LIANG G Q, MU P Q, et al.Improvement of catalytic activity of sorbose dehydrogenase for deoxynivalenol degradation by rational design[J].Food Chemistry, 2023, 423:136274.
[47] SHI Y, XU W, NI D W, et al.Identification and application of a novel deoxynivalenol-degrading enzyme from Youhaiella tibetensis[J].Food Chemistry, 2024, 435:137609.
[48] LIN N N, CHEN J, XU B,et al.The roles of carboxylesterase and CYP isozymes on the in vitro metabolism of T-2 toxin[J].Military Medical Research, 2015, 2:13.
[49] JOHNSEN H, ODDEN E, JOHNSEN B A, et al.Metabolism of T-2 toxin by blood cell carboxylesterases[J].Biochemical Pharmacology, 1988, 37(16):3193-3197.
[50] WU Q H, WANG X, YANG W, et al.Oxidative stress-mediated cytotoxicity and metabolism of T-2 toxin and deoxynivalenol in animals and humans:An update[J].Archives of Toxicology, 2014, 88(7):1309-1326.
[51] 谷晓丹. 漆酶Lac-W广谱高效降解饲料中六种主要霉菌毒素的研究[D].郑州:河南农业大学, 2023.
GU X D.Study on broad-spectrum and efficient degradation of six main mycotoxins in feed by laccase Lac-W[D].Zhengzhou:Henan Agricultural University, 2023.
[52] TSO K H, LUMSANGKUL C, JU J C, et al.The potential of peroxidases extracted from the spent mushroom (Flammulina velutipes) substrate significantly degrade mycotoxin deoxynivalenol[J].Toxins, 2021, 13(1):72.
[53] GAUTÉRIO G V, MALTA D S, REGINATTO L, et al.Use of partially purified peroxidase of agricultural by-product rice bran in deoxynivalenol reduction[J].Journal of Chemical Technology &Biotechnology, 2017, 92(8):1998-2008.
[54] ITO M, SATO I, ISHIZAKA M, et al.Bacterial cytochrome P450 system catabolizing the Fusarium toxin deoxynivalenol[J].Applied and Environmental Microbiology, 2013, 79(5):1619-1628.
[55] KOBAYASHI J, HORIKOSHI T, RYU J C, et al.The cytochrome P-450-dependent hydroxylation of T-2 toxin in various animal species[J].Food and Chemical Toxicology, 1987, 25(7):539-544.
[56] SWANSON S P, HELASZEK C, BUCK W B, et al.The role of intestinal microflora in the metabolism of trichothecene mycotoxins[J].Food and Chemical Toxicology, 1988, 26(10):823-829.
[57] GAO X J, MU P Q, WEN J K, et al.Detoxification of trichothecene mycotoxins by a novel bacterium, Eggerthella sp.DII-9[J].Food and Chemical Toxicology, 2018, 112:310-319.
[58] LI F C, WANG J Q, HUANG L B, et al.Effects of adding Clostridium sp.WJ06 on intestinal morphology and microbial diversity of growing pigs fed with natural deoxynivalenol contaminated wheat[J].Toxins, 2017, 9(12):383.
[59] GAO X J, MU P Q, ZHU X H, et al.Dual function of a novel bacterium, Slackia sp.D-G6:Detoxifying deoxynivalenol and producing the natural estrogen analogue, equol[J].Toxins, 2020, 12(2):85.
[60] HE W J, SHI M M, YANG P, et al.Novel soil bacterium strain Desulfitobacterium sp.PGC-3-9 detoxifies trichothecene mycotoxins in wheat via de-epoxidation under aerobic and anaerobic conditions[J].Toxins, 2020, 12(6):363.
[61] YOUNG J C, ZHOU T, YU H, et al.Degradation of trichothecene mycotoxins by chicken intestinal microbes[J].Food and Chemical Toxicology, 2007, 45(1):136-143.
[62] UENO Y, NAKAYAMA K, ISHII K, et al.Metabolism of T-2 toxin in Curtobacterium sp.strain 114-2[J].Applied and Environmental Microbiology, 1983, 46(1):120-127.
[63] 马妍, 孙长坡, 王峻, 等.T-2毒素降解菌株的筛选、鉴定与降解机制分析[J].食品科学, 2023, 44(22):173-182.
MA Y, SUN C P, WANG J, et al.Screening, identification and action mechanism of T-2 toxin-degrading strains[J].Food Science, 2023, 44(22):173-182.
[64] MATSUSHIMA T, OKAMOTO E, MIYAGAWA E, et al.Deacetylation of diacetoxyscirpenol to 15-acetoxyscirpenol by rumen bacteria[J].The Journal of General and Applied Microbiology, 1996, 42(3):225-234.
[65] FEIZOLLAHI E, ROOPESH M S.Mechanisms of deoxynivalenol (DON) degradation during different treatments:A review[J].Critical Reviews in Food Science and Nutrition, 2022, 62(21):5903-5924.
[66] SCHWARTZ-ZIMMERMANN H E, HAMETNER C, NAGL V, et al.Glucuronidation of deoxynivalenol (DON) by different animal species:Identification of iso-DON glucuronides and iso-deepoxy-DON glucuronides as novel DON metabolites in pigs, rats, mice, and cows[J].Archives of Toxicology, 2017, 91(12):3857-3872.
[67] MICHLMAYR H, MALACHOV A, VARGA E, et al.Biochemical characterization of a recombinant UDP-glucosyltransferase from rice and enzymatic production of deoxynivalenol-3-O-β-D-glucoside[J].Toxins, 2015, 7(7):2685-2700.
A, VARGA E, et al.Biochemical characterization of a recombinant UDP-glucosyltransferase from rice and enzymatic production of deoxynivalenol-3-O-β-D-glucoside[J].Toxins, 2015, 7(7):2685-2700.
[68] POPPENBERGER B, BERTHILLER F, LUCYSHYN D, et al.Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana[J].Journal of Biological Chemistry, 2003, 278(48):47905-47914.
[69] WU X N, MURPHY P, CUNNICK J, et al.Synthesis and characterization of deoxynivalenol glucuronide:Its comparative immunotoxicity with deoxynivalenol[J].Food and Chemical Toxicology, 2007, 45(10):1846-1855.
[70] WELSCH T, HUMPF H U.HT-2 toxin 4-glucuronide as new T-2 toxin metabolite:Enzymatic synthesis, analysis, and species specific formation of T-2 and HT-2 toxin glucuronides by rat, mouse, pig, and human liver microsomes[J].Journal of Agricultural and Food Chemistry, 2012, 60(40):10170-10178.
[71] 孟玲玲. NJA-1菌中DON降解酶基因在毕赤酵母中的表达及其对DON降解效果分析[D].南京:南京农业大学, 2013.
MENG L L.Expression of DON degrading enzyme gene from NJA-1 in Pichia pastoris and its degradation effect on DON[D].Nanjing:Nanjing Agricultural University, 2013.
[72] DOHNAL V, JEZKOVA A, JUN D, et al.Metabolic pathways of T-2 toxin[J].Current Drug Metabolism, 2008, 9(1):77-82.
[73] SHANAKHAT H, MCCORMICK S P, BUSMAN M, et al.Modification of deoxynivalenol by a fungal laccase paired with redox mediator TEMPO[J].Toxins, 2022, 14(8):548.