双歧杆菌(Bifidobacterium)是首批定殖于人体肠道的微生物,与宿主健康密切相关[1]。人体肠道中双歧杆菌的种类和数量随着年龄的增长而变化,是婴儿和成人胃肠道的优势菌群[2]。长双歧杆菌(B.longum)是人体肠道双歧杆菌的优势菌种,占34.30%,在各个年龄段都存在,尤其是在中青年、老人和长寿老人中丰度最高[3]。B. longum具有调节肠道微生态平衡[4]、抑制致病菌生长[5]、调节免疫[6]、缓解便秘[7]、降血脂[8]、预防肥胖和糖尿病[9]、抑制结直肠癌[10]等众多益生特性,且B. longum subsp. longum已被欧洲食品安全局(European Food Safety Agency,EFSA)列入安全资格认定(qualified presumption of safety,QPS)清单[11],因此B.longum已成为全球范围内应用最广泛的益生菌之一。但有些双歧杆菌对氨基糖苷类、四环素类、喹诺酮类和青霉素类抗生素均表现出抗性,且其基因组中也鉴定到相关的抗性基因[12-14]。抗生素抗性基因(antibiotic resistance genes,ARGs)作为一种新型环境污染物,可通过水平基因转移到环境中进行传播和扩散[15]。因此在潜在益生双歧杆菌菌株商业化应用时,应对其抗生素耐药进行安全性评估。
目前人们多聚焦于致病菌耐药性研究,对潜在益生双歧杆菌菌株抗生素耐药性缺乏监测。特别是,不同种双歧杆菌抗生素耐药性判定普遍使用EFSA制定的双歧杆菌属水平微生物折点值(microbiological cut-off values,MCOFFs)[16]。MCOFFs的制定常基于细菌-抗生素组合的最小抑菌浓度(minimal inhibitory concentration,MIC)分布[17],而MIC分布因细菌种类、抗生素类别和抗生素敏感性实验方法的不同而变化[18],因此最好制定细菌种水平MCOFFs。本研究旨在通过测定52株B. longum对6类常见抗生素的MIC值,采用2种经典统计学方法进行B. longum抗生素种特异性初始微生物折点值的制定,以用于区分敏感菌株与获得性抗性菌株,并从分子基因层面进行固有耐药和获得性耐药机制解析。
1 材料与方法
1.1 菌株与试剂
1.1.1 测试菌株
实验所选的52株长双歧杆菌均保藏在江南大学食品生物技术菌种保藏中心,菌株分离信息见表1。
表1 52株长双歧杆菌菌株来源信息表
Table 1 Origin information of 52 B. longum isolates

菌株宿主信息性别年龄地区菌株宿主信息性别年龄地区FJSWXJ2M1女87江苏无锡FHuNa412女3河南博爱JSWXJ24M1女86江苏无锡FGZ19I1M3-新生儿广州合生元FSDLZ61M5-动物粪便山东莱州FGZ17I1M1-新生儿广州合生元FJSNT40M12女83江苏南通C11A10B女108河南博爱FGSZY6M4男1甘肃张掖HUB225男103湖北钟祥FGSZY56M3女9甘肃张掖HUB61男95湖北钟祥FGDLZ19M5女6广东莱州CCFM688女1江苏如皋FJSWXJ37M4女79江苏无锡M203F0227女1西藏当雄FSDLZ56M1女47山东莱州FHeNJZ448女1河南焦作FJSWXJ39M4女84江苏无锡FGZ16I1M5-新生儿广州合生元FGDLZ72M1女70广东莱州FGZ23I1M2-新生儿广州合生元FSDLZ51M1女68山东莱州FXJWS23M7女35新疆乌苏FSDLZ59M1男44山东莱州FXJWS49M9女35新疆乌苏FJSWXJ3M1男83江苏无锡FJSNT19M2女84江苏南通FGDLZ8M1男38广东莱州FSCCD3M1女60四川成都FJSWXJ41M1女88江苏无锡FSCCD5M1女90四川成都HuNa2810---FGSYC28M5男21甘肃永昌FJSWX15M1-新生儿江苏无锡FYNLJ22M3男45云南丽江
续表1
菌株宿主信息性别年龄地区菌株宿主信息性别年龄地区FSDLZ50M2男67山东莱州FSHHK16M1女93上海杨浦FJSNT39M10男66江苏南通FSHHK18M1女82上海杨浦FJSNT32M4女63江苏南通FSHHK25M1女86上海杨浦FJSWXJ20M1男90江苏无锡FSCDJY6M1女63四川都江堰FJSWXJ14M1男78江苏无锡FSCDJY7M2女35四川都江堰FJSWXJ38M1女91江苏无锡FFJNDD11M1女26福建宁德FJSWXJ30M1女85江苏无锡FSCDJY15M2女15四川都江堰FJSWXJ10M2男85江苏无锡FZJTZ1M1女-浙江台州
1.1.2 质控菌株
本研究选用B. longum ATCC 15707作为抗生素敏感性实验的质控菌株,为商业购买菌株。
1.1.3 主要试剂
四环素、红霉素、克林霉素、氨苄青霉素、氯霉素、万古霉素,购于生工生物工程(上海)股份有限公司。
1.1.4 培养基
1.1.4.1 MRS培养基
菌株活化培养基(MRS-Cys)(g):蛋白胨10,牛肉膏10,酵母粉5,葡萄糖20,Na2SO4 2,MgSO4·7H2O 0.5,MnSO4 0.25,柠檬酸氢二铵2,KH2PO4 2.6,吐温1 mL,1 L MRS培养基中添加0.5 g L-半胱氨酸盐酸盐,pH调至6.8。
1.1.4.2 LSM培养基
抗生素敏感性实验用培养基(LSM-Cys):ISO-SENSITEST BROTH (OXOID,货号CM0473B)和MRS培养基按照9∶1的体积比混合为LSM培养基,1 L LSM培养基中添加0.5 g L-半胱氨酸盐酸盐为LSM-Cys培养基。
1.2 仪器与设备
电子天平(ME3002E)、pH计(FE-20),梅特勒-托利多仪器(上海)有限公司;全自动高压蒸汽灭菌锅(SX-500),日本Tomy Digital Biology公司;电热恒温鼓风干燥箱(DGG-9123A),上海森信实验仪器有限公司;超净工作台(ZHJH-C1109B),江苏苏净集团有限公司;紫外可见分光光度计(UV-1800),日本岛津企业管理有限公司;厌氧工作站(Whitley DG250),英国Don Whitley Scientific公司;全波长自动酶标仪(MULTISCAN GO),赛默飞世尔科技(中国)有限公司。
1.3 实验方法
1.3.1 菌株活化
将测试菌株和质控菌株按体积分数1%的接种量接种到MRS-Cys培养基中,置于厌氧工作站,37 ℃培养24~48 h,活化2代。
1.3.2 抗生素敏感性实验
抗生素敏感性实验方法为肉汤微量稀释法,具体操作参照国际标准方法ISO 10932(IDF 223:2010)[19]。
1.3.3 初始微生物折点值的制定
TMCOFFs的制定参照TURNIDGE等[20](T)和KRONVALL[21](K)两种经典统计方法。MIC值经统计分析得MIC累积频数分布表,按照相应的方法说明将频数分布表分别导入ECOFFinder(T法)和Automatic_NRI-MIC_Win_V02beta(K法)2个数据表中。T法计算过程为:先将MIC分布进行以2为底的对数正态分布检验,符合正态分布的MIC分布进行非线性最小二乘法回归拟合,得到3个重要参数:菌株估计值、log2平均值(log2Mean)和log2标准差(log2SD),然后从频数分布最高或高于1个2倍浓度的MIC分布开始迭代拟合,直至包含所有抗生素浓度。当拟合估计值(N)与真值之差最小时,拟合度最佳,此时的MIC为最佳MIC值,基于最佳MIC下的log2平均值和log2标准差计算得到微生物折点值真值。本文选取包含99%野生型菌群的折点值真值向上四舍五入至邻近的2倍抗生素浓度即定义为TMCOFFs[22]。同样地,K法计算过程为:将MIC分布导入数据表后,得到3个重要参数值:log2平均值(log2Mean)、log2标准差(log2SD)和平均值加2个标准差(Mean+2SD);Mean+2SD向上四舍五入至邻近的2倍抗生素浓度即定义为TMCOFFs。
1.3.4 耐药基因的鉴定
将52株测试菌株的基因组序列与抗生素抗性基因数据库(CARD)和可移动耐药基因数据库(ResFinder)比对,以氨基酸序列一致性≥30%和比对覆盖度≥90%为阈值,筛选出推定的抗生素耐药基因[23-24]。
1.3.5 可移动耐药基因序列分析及系统发育树构建
根据ResFinder数据库的鉴定结果提取目的菌株中的可移动耐药基因序列,并在NCBI数据库中下载相应的耐药基因序列,两者通过ClustalW(默认参数)进行多重比对,系统发育树的构建采用MEGA-X。
1.4 数据统计分析
采用HemI 1.0进行可移动耐药基因的热图绘制。
2 结果与分析
2.1 长双歧杆菌抗生素敏感性评估
本研究测定了52株B. longum对四环素、红霉素、克林霉素、氨苄青霉素、氯霉素和万古霉素的MIC值。由表2可知,在MIC分布上,B. longum对四环素的MIC分布范围为0.125~64 μg/mL,菌株FGZ23I1M2的MIC值仅为0.125 μg/mL(表3),而有8株B. longum的MIC高达64 μg/mL。红霉素MIC分布范围为0.032~>8 μg/mL,克林霉素MIC分布范围为0.032~>16 μg/mL,分别覆盖7和6个2倍抗生素浓度梯度。13株B. longum对红霉素和克林霉素的MIC值均超出其测定的最大抗生素浓度范围,并表现出同步抗性。在氨苄青霉素的MIC分布上,绝大多数B. longum的MIC≤4 μg/mL;而在氯霉素MIC分布上,除菌株FJSWXJ2M1的MIC<0.125 μg/mL外(表3),剩下的B. longum菌株的MIC分布仅涵盖4个浓度梯度。而对于万古霉素MIC分布,19%(10/52)B. longum菌株的MIC值明显分离于正常敏感菌群。
表2 52株B. longum对6种抗生素的MIC分布
Table 2 MIC distribution of six antibiotics for 52 B. longum strains
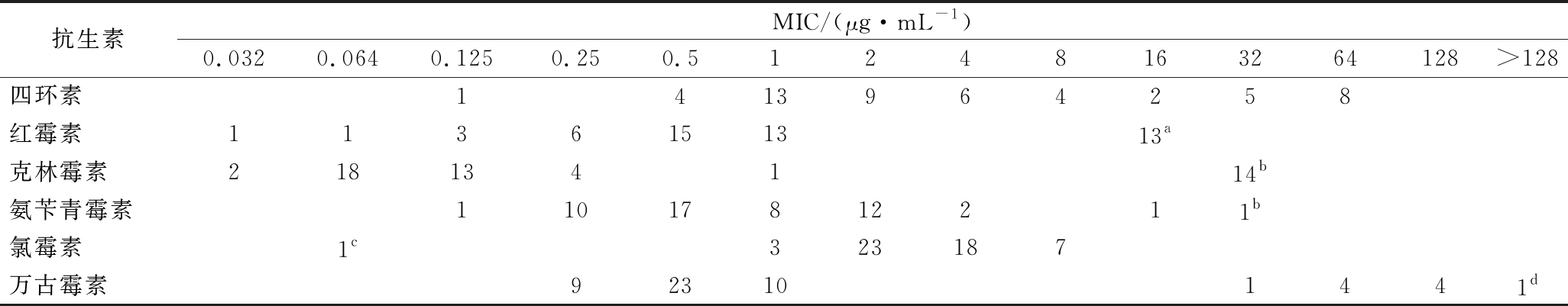
注:a MIC>8 μg/mL; bMIC>16 μg/mL; cMIC<0.125 μg/mL; d MIC>128 μg/mL
抗生素MIC/(μg·mL-1)0.0320.0640.1250.250.51248163264128>128四环素1413964258红霉素1136151313a克林霉素218134114b氨苄青霉素11017812211b氯霉素1c323187万古霉素923101441d
表3 52株B. longum对6种抗生素的MIC分布结果 单位:μg/mL
Table 3 Distribution results of 52 B. longum strains against 6 antibiotics
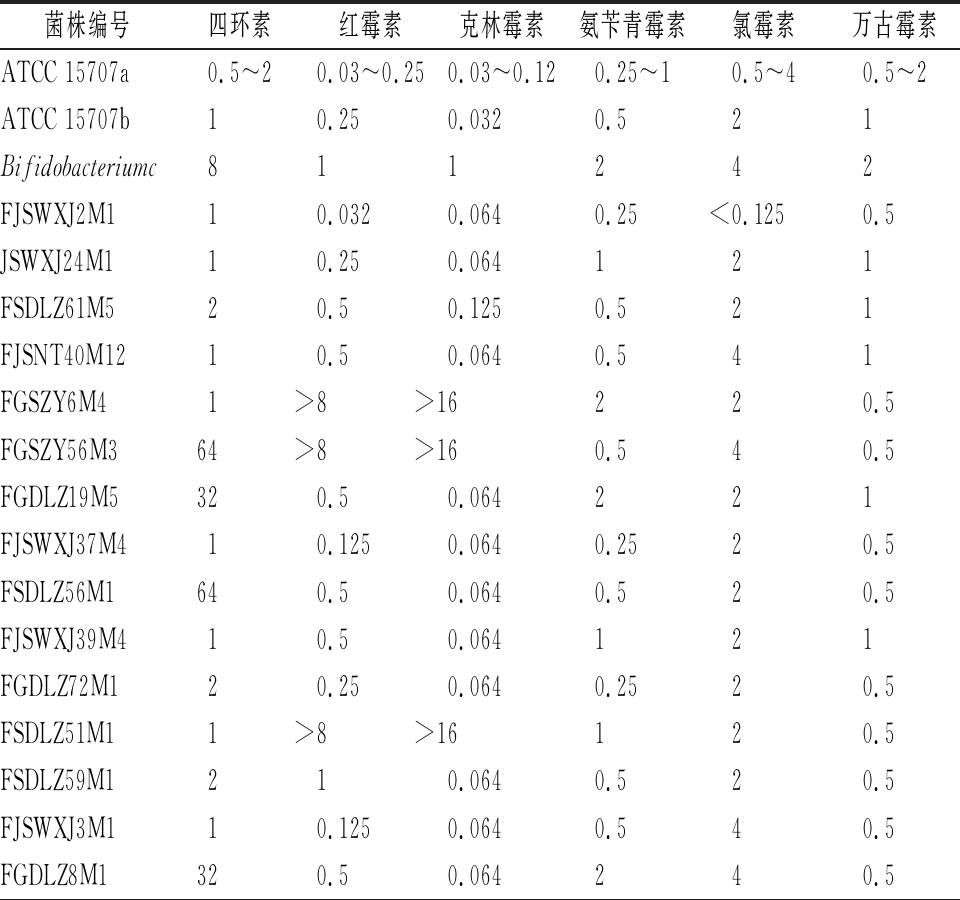
菌株编号四环素红霉素克林霉素氨苄青霉素氯霉素万古霉素ATCC 15707a0.5~20.03~0.250.03~0.120.25~10.5~40.5~2ATCC 15707b10.250.0320.521Bifidobacteriumc811242FJSWXJ2M110.0320.0640.25<0.1250.5JSWXJ24M110.250.064121FSDLZ61M520.50.1250.521FJSNT40M1210.50.0640.541FGSZY6M41>8>16220.5FGSZY56M364>8>160.540.5FGDLZ19M5320.50.064221FJSWXJ37M410.1250.0640.2520.5FSDLZ56M1640.50.0640.520.5FJSWXJ39M410.50.064121FGDLZ72M120.250.0640.2520.5FSDLZ51M11>8>16120.5FSDLZ59M1210.0640.520.5FJSWXJ3M110.1250.0640.540.5FGDLZ8M1320.50.064240.5
续表3
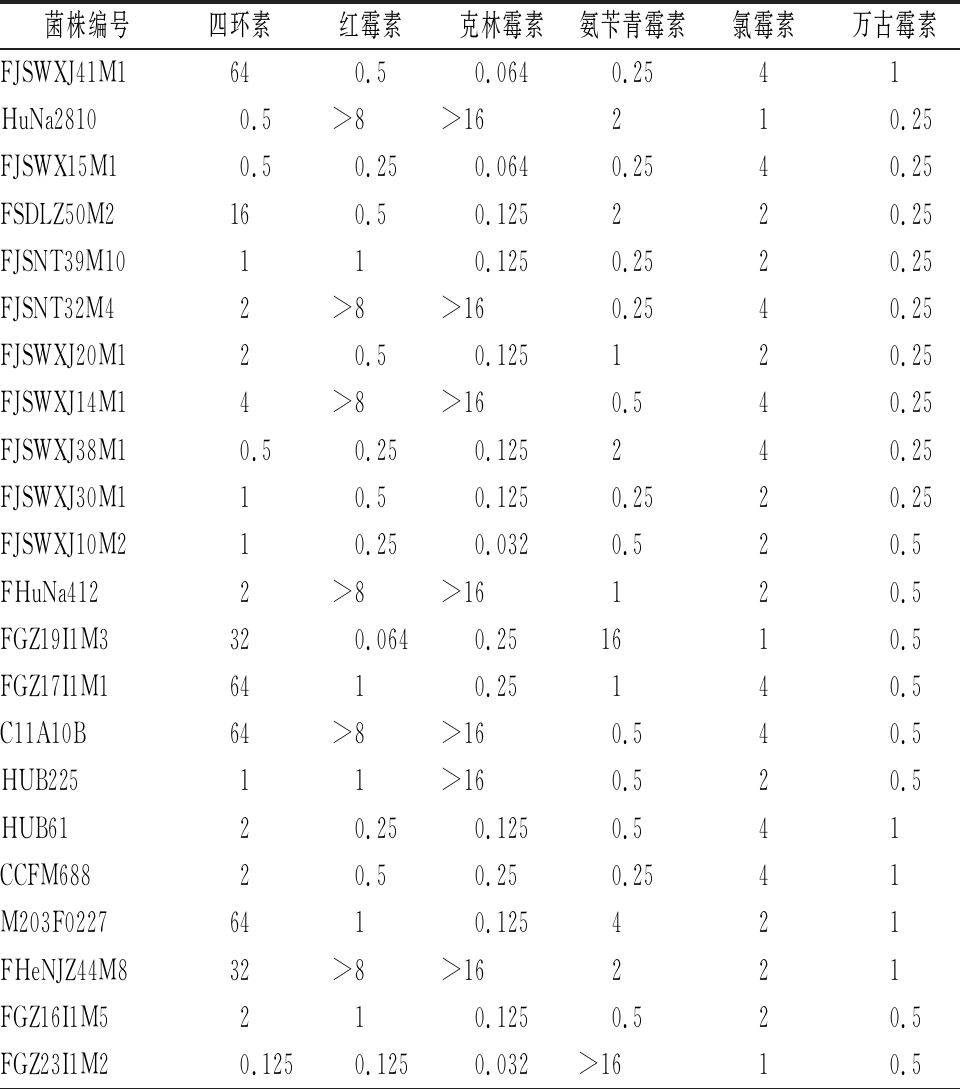
菌株编号四环素红霉素克林霉素氨苄青霉素氯霉素万古霉素FJSWXJ41M1640.50.0640.2541HuNa28100.5>8>16210.25FJSWX15M10.50.250.0640.2540.25FSDLZ50M2160.50.125220.25FJSNT39M10110.1250.2520.25FJSNT32M42>8>160.2540.25FJSWXJ20M120.50.125120.25FJSWXJ14M14>8>160.540.25FJSWXJ38M10.50.250.125240.25FJSWXJ30M110.50.1250.2520.25FJSWXJ10M210.250.0320.520.5FHuNa4122>8>16120.5FGZ19I1M3320.0640.251610.5FGZ17I1M16410.25140.5C11A10B64>8>160.540.5HUB22511>160.520.5HUB6120.250.1250.541CCFM68820.50.250.2541M203F02276410.125421FHeNJZ44M832>8>16221FGZ16I1M5210.1250.520.5FGZ23I1M20.1250.1250.032>1610.5
续表3
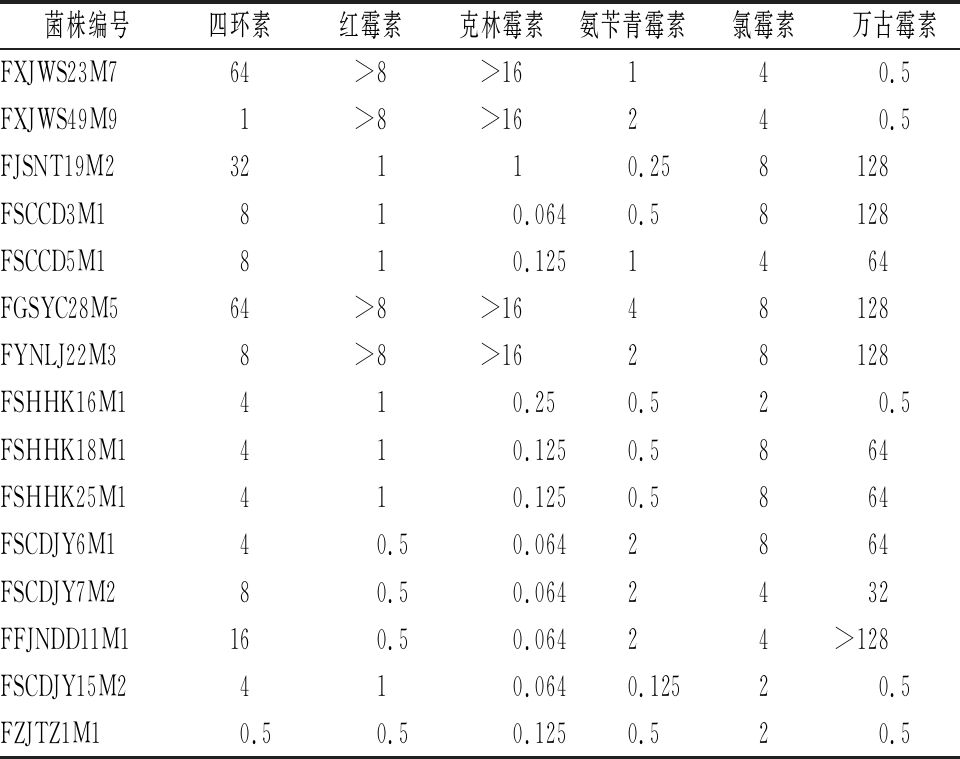
注:a,质控菌株的质控范围;b,质控菌株的实际测定值;c,EFSA制定的双歧杆菌属水平微生物临界值
菌株编号四环素红霉素克林霉素氨苄青霉素氯霉素万古霉素FXJWS23M764>8>16140.5FXJWS49M91>8>16240.5FJSNT19M232110.258128FSCCD3M1810.0640.58128FSCCD5M1810.1251464FGSYC28M564>8>1648128FYNLJ22M38>8>1628128FSHHK16M1410.250.520.5FSHHK18M1410.1250.5864FSHHK25M1410.1250.5864FSCDJY6M140.50.0642864FSCDJY7M280.50.0642432FFJNDD11M1160.50.06424>128FSCDJY15M2410.0640.12520.5FZJTZ1M10.50.50.1250.520.5
2.2 长双歧杆菌抗生素耐药性微生物折点值制定
本文基于52株B. longum对6种抗生素的MIC频数分布,采用Turnidge(T)和Kronvall(K)两种经典统计学方法,进行B. longum种特异性TMCOFFs的制定及耐药率比较分析,并与已制定的双歧杆菌属水平折点值进行比较。表4为2种统计方法计算过程中得到的重要参数值:log2Mean、log2SD和基于前面2个参数计算得到的折点值真值,折点值真值向上四舍五入至邻近的2倍抗生素浓度,即为TMCOFFs。由表5可知,采用T法制定的B. longum对四环素、红霉素、克林霉素、氨苄青霉素、氯霉素和万古霉素的TMCOFFs依次为8、8、0.25、8、8和2 μg/mL;而基于K法计算得到的B. longum对这6种抗生素的TMCOFFs依次为8、8、0.25、2、8和2 μg/mL。2种不同统计方法计算结果相比较发现:除氨苄青霉素TMCOFFs相差2个抗生素浓度外,T法与K法计算得到的B. longum与剩下5种抗生素组合的TMCOFFs完全相等,这表明2种统计方法计算结果的一致性非常高。因此本文初步制定的B. longum对给定抗生素的种特异性微生物折点值是可靠的。
表4 基于Turnidge和Kronvall方法下长双歧杆菌折点值的估计
Table 4 Estimates of cut-off values for B. longum based on Turnidge and Kronvall methods
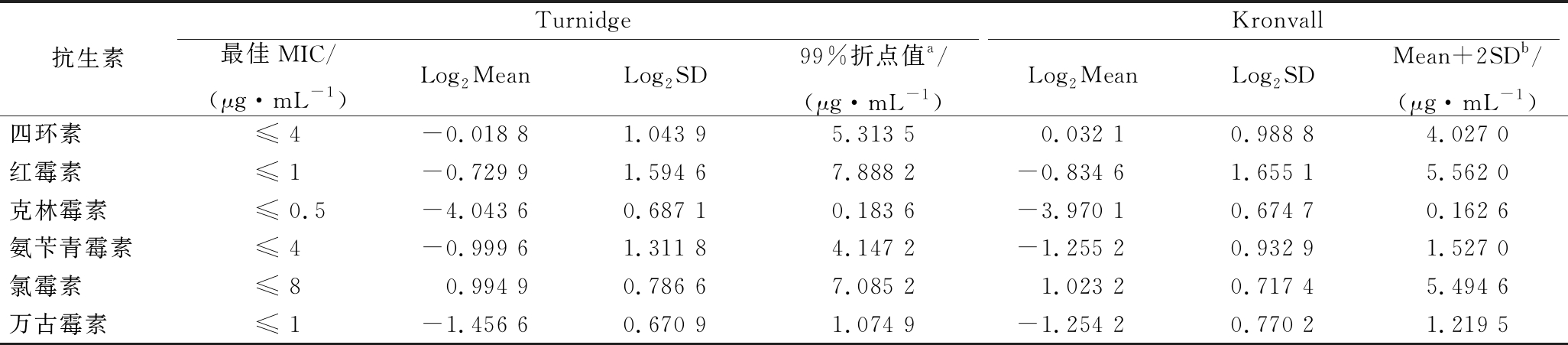
注:a,采用Turnidge方法计算得到包含99%野生型菌群的折点值真值;b,采用Kronvall方法计算的折点值真值
抗生素TurnidgeKronvall最佳MIC/(μg·mL-1)Log2MeanLog2SD99%折点值a/(μg·mL-1)Log2MeanLog2SDMean+2SDb/(μg·mL-1)四环素≤ 4-0.018 81.043 95.313 50.032 10.988 84.027 0红霉素≤ 1-0.729 91.594 67.888 2-0.834 61.655 15.562 0克林霉素≤ 0.5-4.043 60.687 10.183 6-3.970 10.674 70.162 6氨苄青霉素≤ 4-0.999 61.311 84.147 2-1.255 20.932 91.527 0氯霉素≤ 80.994 90.786 67.085 21.023 20.717 45.494 6万古霉素≤ 1-1.456 60.670 91.074 9-1.254 20.770 21.219 5
表5 52株B. longum的初始微生物折点值和耐药率统计结果
Table 5 The tentative microbiological cut-off values, and the statistical results of resistance rates of 52 B. longum strains

注:aEFSA制定的双歧杆菌属水平微生物折点值;b采用Turnidge方法制定的B. longum抗生素初始微生物折点值;c采用Kronvall方法制定的B. longum抗生素初始微生物折点值
抗生素EFSATurnidgeKronvallTMCOFFsa/(μg·mL-1)耐药菌株数(耐药率/%)TMCOFFsb/(μg·mL-1)耐药菌株数(耐药率/%)TMCOFFsc/(μg·mL-1)耐药菌株数(耐药率/%)四环素815(28.85)815(28.85)815(28.85)红霉素113(25)813(25)813(25)克林霉素114(26.93)0.2515(28.85)0.2515(28.85)氨苄青霉素24(7.69)82(3.85)24(7.69)氯霉素47(13.46)8080万古霉素210(19.23)210(19.23)210(19.23)
种特异性折点值与EFSA属水平折点值相比较发现:种水平和属水平折点值之间既存在一致性又存在差异性;相较于属水平折点值,基于种特异性折点值B. longum对四环素、红霉素、克林霉素和万古霉素耐药率水平相当,为中度抗性;氨苄青霉素耐药率水平均较低;而氯霉素耐药率由13.46%降为0。
2.3 种特异性微生物折点值下长双歧杆菌多重耐药性分析
基于种特异性微生物折点值进行B. longum对给定抗生素表型抗性的鉴定,若菌株对3类或3类以上抗生素表现出抗性时,即定义为多重耐药[25]。表6为52株长双歧杆菌对6种抗生素的多重耐药性统计结果,由表6可知,21(40.4%)株菌为野生型,14株菌对1种抗生素有抗性,11株菌对2种抗生素具有抗性。5株菌对3种抗生素具有抗性,1株菌对5种抗生素具有抗性。6(11.5%)株菌为多重耐药菌,其中FGSYC28M5是耐药谱最广的菌株,耐药谱为四环素-红霉素-克林霉素-氨苄青霉素-万古霉素(表3)。总体来看,基于B. longum种特异性微生物折点值对52株分离株与给定6种抗生素的多重耐药分析,发现其多重耐药情况较轻,为今后长双歧杆菌的安全性应用提供参考。
表6 52株B. longum的多重耐药性结果
Table 6 Multi-drug resistance results of 52B. longum strains
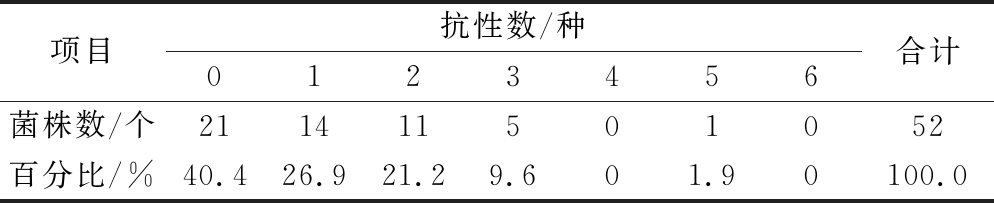
注:B. longum对四环素、红霉素、克林霉素、氨苄青霉素、氯霉素和万古霉素的耐药折点值选用K法计算得到的种特异性折点值
项目抗性数/种0123456合计菌株数/个211411501052百分比/%40.426.921.29.601.90100.0
2.4 长双歧杆菌抗生素耐药性基因分析
表7为52株B. longum菌株基因组与CARD数据库比对鉴定的表型抗性相关的耐药基因。由表7可知,52株测试菌株均比对到了四环素耐药基因rpsJ和tetB(P),红霉素耐药基因macB,克林霉素耐药基因lmrB、lmrC和lmrD,而B. longum对这3种抗生素的表型耐药率依次为28.85%、25%和28.85%;故推测B. longum对这3种抗生素耐受可能由其他新的耐药基因介导或需多个耐药基因共同作用。但仅有17株菌株比对到四环素抗性基因tet(W),且其序列一致性高达97%,其中13株菌株为四环素耐药菌株;而且通过可移动耐药基因数据库ResFinder鉴定发现,13株四环素耐药菌株中,可移动耐药基因tet(W)的检出率高达92.31%(12/13)(图1),故推测基因tet(W)可提高长双歧杆菌四环素耐受性,且该基因具有水平基因转移至其他致病菌的风险。有13株B. longum菌株同时表现出红霉素和克林霉素抗性,其中10株菌株基因组中均存在可移动抗性基因erm(X)(图1),推测B. longum对红霉素和克林霉素共同抗性是由可移动耐药基因erm(X)介导,这与MARTíNEZ等[26]的研究结果一致;而菌株FJSWXJ41M1和FSDLZ50M2均含有erm(X),但其对红霉素和克林霉素均敏感,因此推测其基因erm(X)可能为沉默基因。所有菌株均未比对到β-内酰胺类抗性基因,这与B. longum对氨苄青霉素表型敏感率吻合程度高达92.31%。51株菌均比对到了多重耐药基因CIPa,1株菌比对到了clbB,而B. longum的多重耐药率仅为11.5%,故推测CIPa和clbB可能为沉默基因或需与多个耐药基因共同作用。B. longum基因组中比对到的糖肽类抗性基因有vanB、vanD、vanRO、vanSO、vanRI和vanRA;WEI等[27]在B. longum JDM301中鉴定到推定的万古霉素抗性基因有vanD、vanU、vanH、vanSB、vanSD5和vanSc3,而本文中10株万古霉素耐药菌株基因组中均只存在vanD基因,故推测该基因是部分B. longum糖肽类抗生素耐受的关键抗性基因。
表7 52株B. longum中表型抗性相关耐药基因的鉴定结果
Table 7 The identification results of antibiotic resistance genes related to phenotype of 52 B. longum strains
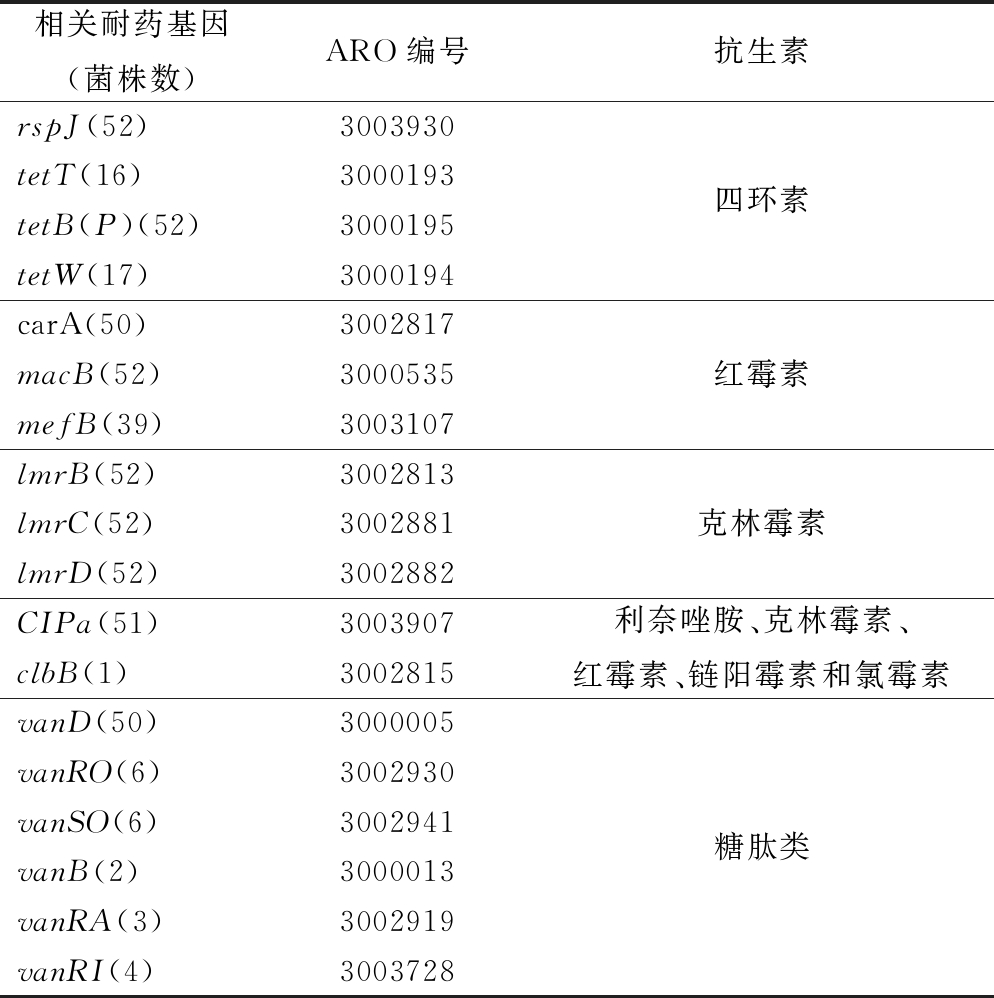
注:没有检测到β-内酰胺类相关的抗生素抗性基因
相关耐药基因(菌株数)ARO编号抗生素rspJ(52)3003930tetT(16)3000193tetB(P)(52)3000195tetW(17)3000194四环素carA(50)3002817macB(52)3000535mefB(39)3003107红霉素lmrB(52)3002813lmrC(52)3002881lmrD(52)3002882克林霉素CIPa(51)3003907clbB(1)3002815利奈唑胺、克林霉素、红霉素、链阳霉素和氯霉素vanD(50)3000005vanRO(6)3002930vanSO(6)3002941vanB(2)3000013vanRA(3)3002919vanRI(4)3003728糖肽类

图1 52株长双歧杆菌菌株基因组中可移动耐药基因tet(W)和erm(X)的分布结果
Fig.1 The distribution results of mobile antibiotic resistance genes tet(W) and erm(X) of 52 B. longum strains genome
2.5 可移动耐药基因tet(W)和erm(X)的系统进化分析
将含有可移动耐药基因的B. longum菌株的tet(W)和erm(X)耐药基因序列与NCBI数据库中下载的11个tet(W)和4个erm(X)基因序列进行比对分析,并采用MEGA-X中的Neighbor-Joining法构建系统发育树,见图2和图3。
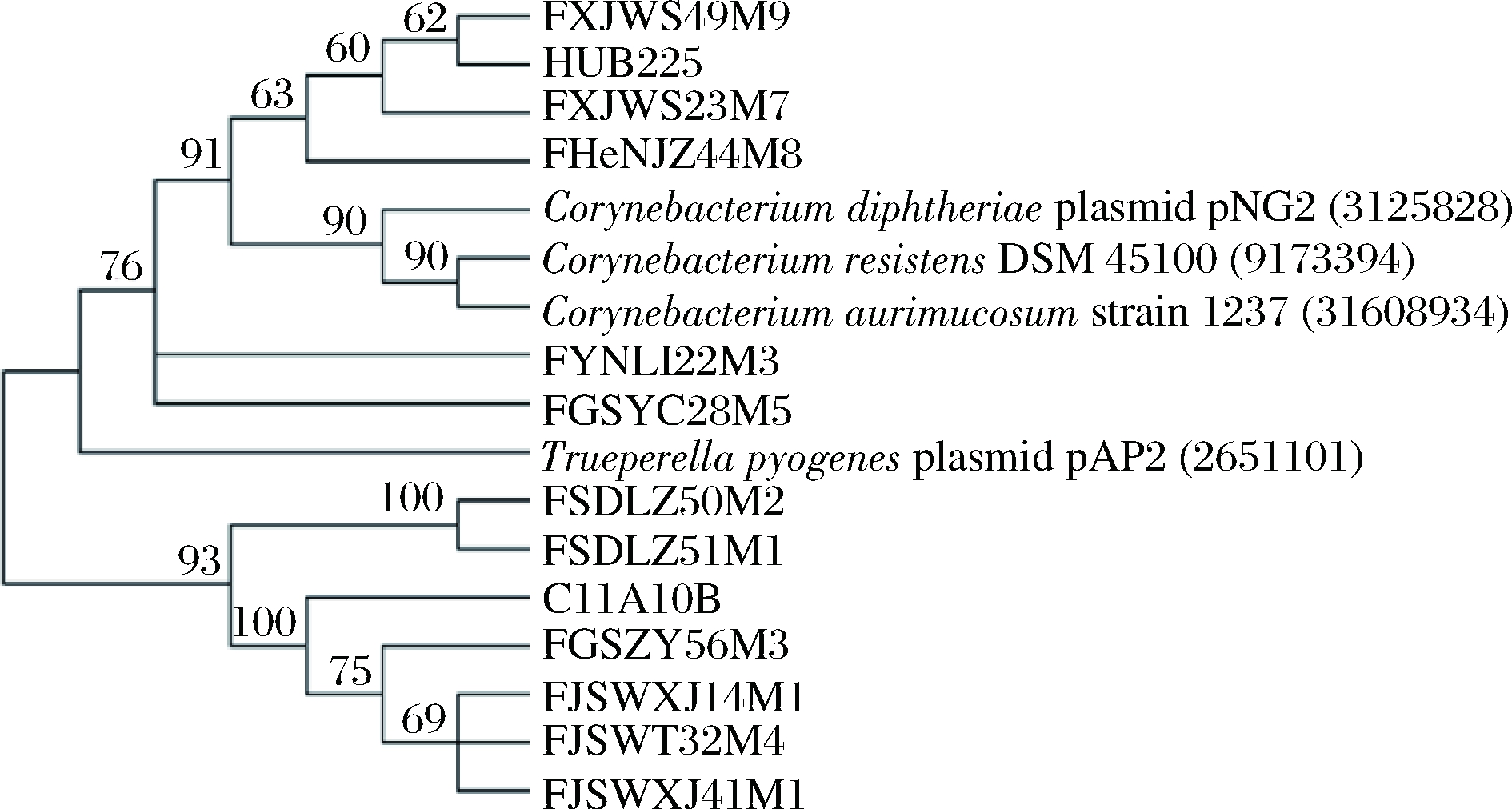
图3 长双歧杆菌中可移动耐药基因erm(X)的系统进化树
Fig.3 The phylogenetic tree of mobile resistance genes oferm(X) of B. longum
注:黑体加粗的菌株为NCBI下载序列;节点上的数字为1 000次重复的支持率

图2 长双歧杆菌中可移动耐药基因tet(W)的系统进化树
Fig.2 The phylogenetic tree of mobile resistance genes oftet(W) of B. longum
由图2可知,15株B. longum菌株分布在2个分支上,其中的13株B. longum菌株与NCBI库下载的长双歧杆菌长亚种(Bifidobacterium longum subsp. longum)和婴儿亚种(B. longum bv. infantis)、两歧双歧杆菌(B. bifidum)、假小链双歧杆菌(B. pseudocatenulatum)、动物双歧杆菌乳亚种(B. animalis subsp. lactis)、杰氏棒杆菌(Corynebacterium jeikeium)和Parasutterella excrementihominis在同一分支上,其序列相似度高,推测可能来源于同一耐药基因;而菌株FGDLZ19M5、FGDLZ8M1和FJSNT32M4与NCBI下载的短双歧杆菌(B. breve)和长双歧杆菌(B. longum)位于另一分支上,且各基因间的进化距离较远,可能来源于不同供体。
由图3可知,13株B. longum菌株分布在2个大分支上,说明不同B. longum菌株的erm(X)基因序列具有显著的变异。其中6株B. longum菌株FXJWS49M9、HUB225、FXJWS23M7、FHeNJZ44M8、FYNLJ22M3和FGSYC28M5与NCBI数据库下载的3种棒状杆菌(Corynebacterium)菌株和化脓隐秘杆菌(Trueperella pyogenes)菌株的erm(X)基因序列同源性较高,推测B. longum菌株中的erm(X)基因可能是由其他细菌水平转移耐药基因所获得。此外,第2个分支上分离自同一地区菌株FSDLZ50M2、FSDLZ51M1、FJSWXJ41M1、FJSNT32M4、FJSWXJ41M1的erm(X)基因序列相似度达到100%。
3 结论
本研究基于长双歧杆菌与6种抗生素组合的MIC分布,初步制定B. longum对四环素、红霉素、克林霉素、氨苄青霉素、氯霉素和万古霉素的种特异性微生物折点值分别为8、8、0.25、8/2(T/K)、8和2 μg/mL,其可用于区分敏感菌株与获得性抗性菌株。25%的B. longum菌株同时表现出红霉素和克林霉素抗性,其抗性是由可移动耐药基因erm(X)介导;可移动耐药基因基因tet(W)介导B. longum四环素表型抗性。系统进化分析表明长双歧杆菌耐药基因tet(W)和erm(X)来源于不同宿主,同一地区分离菌株的耐药基因具有高度同源性。益生菌菌株的耐药性判定关系到食品安全,本文通过制定长双歧杆菌的耐药性折点值,对长双歧杆菌在食品中的安全性应用具有指导价值。
[1] O′CALLAGHAN A,VAN S D.Bifidobacteria and their role as members of the human gut microbiota[J].Frontiers in Microbiology,2016,7:925.
[2] 丁喜顺.长双歧杆菌细胞壁完整肽聚糖的分离及其免疫、抑瘤活性的研究[D].呼和浩特:内蒙古农业大学,2009.
[3] 闫爽.长双歧杆菌遗传与表型多样性及其与免疫调节功能的相关性研究[D].无锡:江南大学,2019.
[4] JANINA M,ANNETTE H,CLAUDIUS M,et al.Efficacy of Bifidobacterium longum, B. infantis and Lactobacillus acidophilus probiotics to prevent gut dysbiosis in preterm infants of 28+0-32+6 weeks of gestation: a randomised, placebo-controlled, double-blind, multicentre trial: the PRIMAL clinical study protocol[J].BMJ Open,2019,9(11):e032617.
[5] INTURRI R,TROVATO L,VOLTI G L,et al.In vitro inhibitory activity of Bifidobacterium longum BB536 and Lactobacillus rhamnosus HN001 alone or in combination against bacterial and Candida reference strains and clinical isolates[J].Heliyon,2019,5(11):e02891.
[6] MIJOO C,YUNJUNG L,NA-KYOUNG L,et al.Immunomodulatory effects by Bifidobacterium longum KACC 91563 in mouse splenocytes and macrophages[J].Journal of Microbiology and Biotechnology,2019,29(11):1 739-1 744.
[7] 丁圣,蒋菁莉,刘松玲,等.长双歧杆菌BBMN68对便秘模型小鼠的通便作用[J].食品科学,2011,32(3):195-198.
[8] XIAO J Z,KONDO S,TAKAHASHI N,et al.Effects of milk products fermented by Bifidobacterium longum on blood lipids in rats and healthy adult male volunteers[J].Journal of Dairy Science,2003,86(7):2 452-2 461.
[9] OTHMAN M B,SAKAMOTO K.Effect of inactivated Bifidobacterium longum intake on obese diabetes model mice (TSOD)[J].Food Research International,2020,129(108 792).
[10] FAHMY C A,GAMAL-ELDEEN A M, EL-HUSSIENY E A,et al.Bifidobacterium longum suppresses murine colorectal cancer through the modulation of oncomi.Rs and tumor suppressor miRNAs[J].Nutrition and Cancer,2019,71(4):688-700.
[11] EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards).Scientific opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2013 update)[J].EFSA Journal,2013,11(11):3 449.
[12] 潘伟好,李平兰,孙承虎.双歧杆菌的药敏性及其质粒DNA的检测[J].食品科学,2005,26(6):81-86.
[13] DURANTI S,LUGLI G A,MANCABELLI L,et al.Prevalence of antibiotic resistance genes among human gut-derived bifidobacteria[J].Applied and Environmental Microbiology,2016,83(3):e02 894-16.
[14] FOUHY F,O’CONNELL M M,FITZGERALD G F,et al.In silico assigned resistance genes confer Bifidobacterium with partial resistance to aminoglycosides but not to β-lactams[J].PloS One,2013,8(12):e82653.
[15] 罗义,周启星.抗生素抗性基因(ARGs)——一种新型环境污染物[J].环境科学学报,2008,28(8):1 499-1 505.
[16] European Food Safety Authority (EFSA).Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance[J].EFSA Journal,2012,10(6):2 740.
[17] European Committee on Antimicrobial Susceptibility Testing.MIC distributions and epidemiological cut-off value (ECOFF) setting[P].EUCAST SOP 10.0,2017.http://www.eucast.org.
[18] KLARE I, KONSTABEL C, WERNER G,et al.Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use[J].Journal of Antimicrobial Chemotherapy,2007,59(5):900-912.
[19] International Stardand Organization (ISO).Milk and milk products-determination of the minimal inhibitory concentration (MIC) of antibiotics applicable to bifidobacteria and non-enterococcal lactic acid bacteria (LAB)[S].ISO,2010.
[20] TURNIDGE J,KAHLMETER G,KRONVALL G.Statistical characterisation of bacterial wild-type MIC value distributions and the determination of microbiological cut-off values[J].Clinical Microbiology and Infection,2006,12(5):418-425.
[21] KRONVALL G.Normalized resistance interpretation as a tool for establishing epidemiological MIC susceptibility breakpoints[J].Journal of Clinical Microbiology,2010,48(12):4 445-4 452.
[22] European Committee on Antimicrobial Susceptibility Testing (EUCAST). EUCAST general consultation on “considerations in the numerical estimation of epidemiological cutoff (ECOFF) values”[J].EUCAST,2018.
[23] CAMPEDELLI I,MATHUR H,SALVETTI E,et al.Genus-wide assessment of antibiotic resistance in Lactobacillus spp.[J].Applied and Environmental Microbiology,2019,85(1):e01 738-18.
[24] HU Y,YANG X,LI J,et al.The bacterial mobile resistome transfer network connecting the animal and human microbiomes[J].Applied and Environmental Microbiology,2016,82(22):6 672-6 681.
[25] 迟星云,朱晓莉,王虹,等.重症监护病房多重耐药菌检出及药物敏感性[J].中华医院感染学杂志,2019,29(24):3 729-3 733.
[26] MARTíNEZ N,LUQUE R,MILANI C,et al.A gene homologous to rRNA methylase genes confers erythromycin and clindamycin resistance in Bifidobacterium breve[J]. Applied and Environmental Microbiology,2018,84(10):e02 888-17.
[27] WEI Y X.Safety assessment of Bifidobacterium longumJDM301 based on complete genome sequences[J].World Journal of Gastroenterology,2012,18(5):479-488.