霉菌毒素是霉菌的次级代谢产物,目前已知的有400余种,主要由曲霉属、镰孢属和青霉属真菌产生[1]。对人和动物危害较大的有黄曲霉毒素(Aflatoxin)、赭曲霉毒素(Ochratoxin)、伏马毒素(Fumonisin)、玉米赤霉烯酮(Zearalenone)和单端孢霉烯毒素(Trichothecene)等几类,主要表现在肝脏毒性、肾脏毒性、致癌性、致畸性、致突变和免疫抑制等方面[2-3]。
霉菌毒素对农作物污染范围广且不容易在收获前有效清除[4],因而研究脱除霉菌毒素的方法十分必要。目前对脱除霉菌毒素的研究,主要分为物理法、化学法和生物法。物理法常用的手段是清洗、吸附、辐射和热处理,比较新颖的方式有电解水处理、脉冲光束和冷等离子体处理[5],但这些方法对设备要求高,能源消耗大,脱毒过程会对营养成分造成破坏,一般只适合霉变程度轻且颗粒大的物料。化学法可分为酸、碱处理、氨处理以及强氧化剂处理,利用霉菌毒素在强酸强碱或者氧化环境中结构不稳定,将之破坏,但化学反应降解毒素容易产生各种副产物,且化学试剂残留会造成二次污染[6]。不同于物理和化学脱毒法,生物法利用各种微生物脱除霉菌毒素,具有反应条件温和、脱毒能耗小、环境友好、营养物质破坏少、特异性强等优点[7]。
1 生物脱毒
1.1 生物吸附脱毒
微生物或微生物材料与毒素结合形成吸附复合物,再由过滤等方式除去,通常适合于液体物料或者溶液体系。
HASKARD等[8]研究了乳酸菌与黄曲霉毒素B1(aflatoxin B1, AFB1)在培养基中的结合效果,证明毒素可以被乳酸菌吸附,在4~37 ℃ 和pH 2~10范围内没有明显的解吸现象,但所形成的结合仍是可逆的。此后陆续有报道利用乳酸菌除去小麦中的脱氧雪腐烯醇(deoxynivalenol, DON)、T-2和HT-2等毒素[9],在食品发酵过程中结合伏马毒素B1和B2(fumonisin B1, FB1、fumonisin B2, FB2),并降低其生物利用度和毒性[10];也可以在牛奶中吸附结合黄曲霉毒素M1(AFM1)[11],吸附效果在34.0%~78.9%范围内不等(表1)。研究也证明酵母可以吸附玉米赤霉烯酮(zearalenone, ZEA)、赭曲霉毒素A(ochratoxin A,OTA)和AFB1、AFM1等多种毒素[12]。CHLEBICZ等[13]研究了12种乳酸菌和6种酵母对霉菌毒素的吸附能力,发现它们对伏马毒素的吸附能力最强,对DON的吸附最弱。随后,人们开始从食物中筛选分离不同的菌种进行研究,如利用Kefiri(主要为乳酸菌和醋酸杆菌)对AFB1、ZEA和OTA进行吸附,吸附量为82%~100%,并证明了在胃肠道中比较稳定,不发生解吸[14];从酸奶中分离的一种副干酪乳酸菌LHZ-1可以吸附脱除DON,且活细胞比灭活细胞效果更好,脱除率达到40.7%[15]。酿酒酵母可以在5 min内去除葡萄汁中90%的OTA[16]。相同条件下,几种乳杆菌与酵母对ZEA的吸附效果进行了比较,吸附效率最佳菌种为巴氏乳杆菌(53.3%)和酿酒酵母(36.5%)[17]。此外,研究者们不仅关注微生物与毒素的吸附率,还借助先进的设备观察与毒素的结合位点。副干酪乳杆菌可以与AFB1特异结合,在扫描电镜下观察到其细胞壁表面空间结构发生变化[18],在胃肠道环境下观察酵母与OTA的结合比在培养基中更加稳定[19]。利用激光共聚焦显微镜观察发现植物乳杆菌与伏马毒素的官能团结合,与FB1的结合最有效[20],吸附部位为细胞壁上的多糖和蛋白。酵母葡甘露聚糖和β-葡聚糖也可以有效地减轻黄曲霉毒素的毒性,基本消除饲料中毒素对肉鸡免疫功能和肝肾功能的不良影响[21]。此外,双歧杆菌属的微生物和芽孢杆菌等也可以通过生物吸附脱除AFB1、AFM1、OTA等霉菌毒素[22-24](表1)。
表1 各种微生物对霉菌毒素的吸附作用
Table 1 Adsorption of mycotoxins using microorganisms
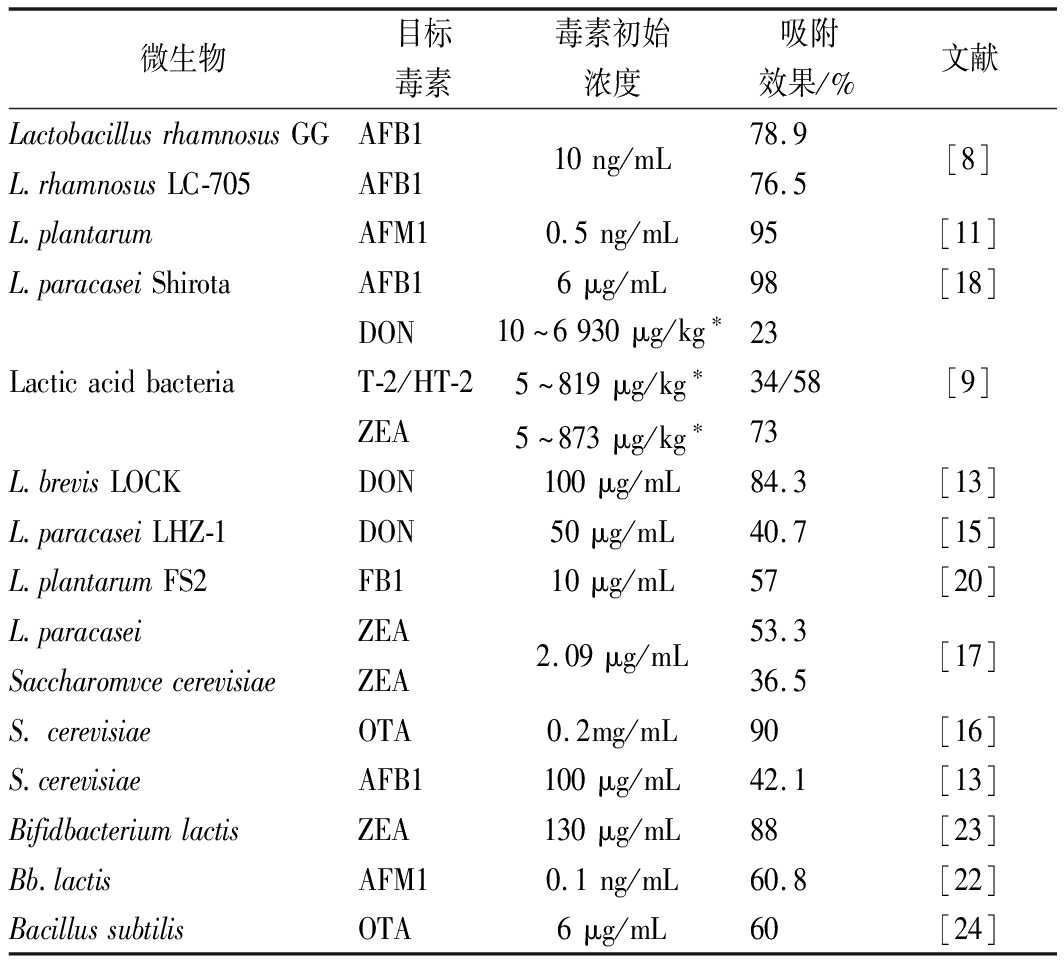
微生物目标毒素毒素初始浓度吸附效果/%文献LactobacillusrhamnosusGGL.rhamnosusLC-705AFB1AFB110ng/mL78.976.5[8]L.plantarumAFM10.5ng/mL95[11]L.paracaseiShirotaAFB16μg/mL98[18]LacticacidbacteriaDONT-2/HT-2ZEA10~6930μg/kg∗5~819μg/kg∗5~873μg/kg∗2334/5873[9]L.brevisLOCKDON100μg/mL84.3[13]L.paracaseiLHZ-1DON50μg/mL40.7[15]L.plantarumFS2FB110μg/mL57[20]L.paracaseiSaccharomvcecerevisiaeZEAZEA2.09μg/mL53.336.5[17]S.cerevisiaeOTA0.2mg/mL90[16]S.cerevisiaeAFB1100μg/mL42.1[13]BifidbacteriumlactisZEA130μg/mL88[23]Bb.lactisAFM10.1ng/mL60.8[22]BacillussubtilisOTA6μg/mL60[24]
注:* 样品为被霉菌毒素污染的小麦,不同批次间含量相差较大,吸附率为平均值。其余样品与毒素的吸附过程均在PBS溶液或液体培养基中进行
1.2 生物降解脱毒
降解是生物法脱除霉菌毒素最重要的一个方面,利用微生物或酶对毒素特异性降解,得到无毒或低毒的降解产物。微生物分泌的酶多种多样,对各种毒素降解方式也不相同,涉及到乙酰化、糖基化、开环、脱羧等反应[1]。
酵母不仅可以吸附AFB1,也可对其进行降解,尤其与枯草芽孢杆菌和干酪乳杆菌混合发酵可减少85%以上的AFB1(68.55 ng/mL)[25]。鱼肠道中分离的一株枯草芽孢杆菌,可以降解81.5%的AFB1(100 ng/mL),还能降解AFM1和AFG1[26],但降解产物复杂。最近研究发现,红球菌[27]和拟香味菌[28]都可以有效地对黄曲霉毒素进行降解(表2)。
由于产物多样,关于黄曲霉毒素的降解机理研究大多为推测。其中,假蜜环菌被发现可以分泌氧化酶降解AFB1结构中的双呋喃环,通过薄层层析和动物实验证明了经过酶处理后的AFB1被完全降解且毒性降低(图1)[29]。
表2 各种微生物降解霉菌毒素的效果
Table 2 Degradation of mycotoxins using microorganisms

微生物目标毒素毒素初始浓度降解率/%机理或途径文献ArmillariellatabescensAFB116μmol/L100打开双呋喃环[29](Pleurotusostreatusetal)AFB11.4μg/mL9.32~52.36真菌漆酶[30]L.casei,B.subtilisandPichiaanomalaAFB168.55ng/mL85~95还原不饱和键[25]MyroidesodoratimimusAFB1100ng/mL93.82胞外酶[28]B.subtilisAFB1(AFM1和AFG1)100ng/mL81.5胞外酶[26]RhodococcuserythropolisAFB11μg/mL>84氧化酶[27]TrichosporonZEA95水解酶[38]B.velezensisA2ZEA7.45μg/mL100-[33]B.pumilusES-21ZEA17.9μg/mL97.5水解、脱羧[34]B.licheniformisCK1ZEA2μg/mL>95.8-[35]B.subtilis168B.nattoZEAZEA20ng/mL81100水解、脱羧[39]PseudomonaFB110μg/mL43~100脱羧基、脱氨基[42]EubacteriumcallanderiOTA0.2μg/mL95-[45]PediococcusparvulusAcinetobacterYarrowialipolyticaOTAOTAOTA1μg/mL90-97.2羧肽酶[47]B.subtilisOTA6μg/mL47.1~97.6水解内酯环[24]R.stoloniferOTA7.5μg/mL90蛋白酶[44]L.sakeiT-2-58-[9]L.rhamnosusDON10μg/mL100去环氧化[52]EggerthellaDONT-2/HT-220μg/mL100去环氧化[49]DesulfitoDONNIV100μg/mL-环氧氧化为羰基[51]

图1 假密环菌降解AFB1中双呋喃环脱毒
Fig.1 Detoxification AFB1 by Armillariella tabescens through difuran ring degradation
ALBERTS等[30]研究了54株真菌对黄曲霉毒素B1的降解作用,发现培养基中漆酶活性与AFB1降解率呈现显著相关,最佳条件下隔孢伏革菌SCC0152可以降解52.36%的AFB1(1.4 μg/mL),利用鼠伤寒沙门氏菌进行致突变毒性研究,证明降解产物的毒性明显小于AFB1。黑曲霉、根霉以及不产生AFs的黄曲霉对于AFB1的降解结果表明,它们通过还原AFB1的酮羰基或双呋喃环上的双键进行解毒[31](图2)。
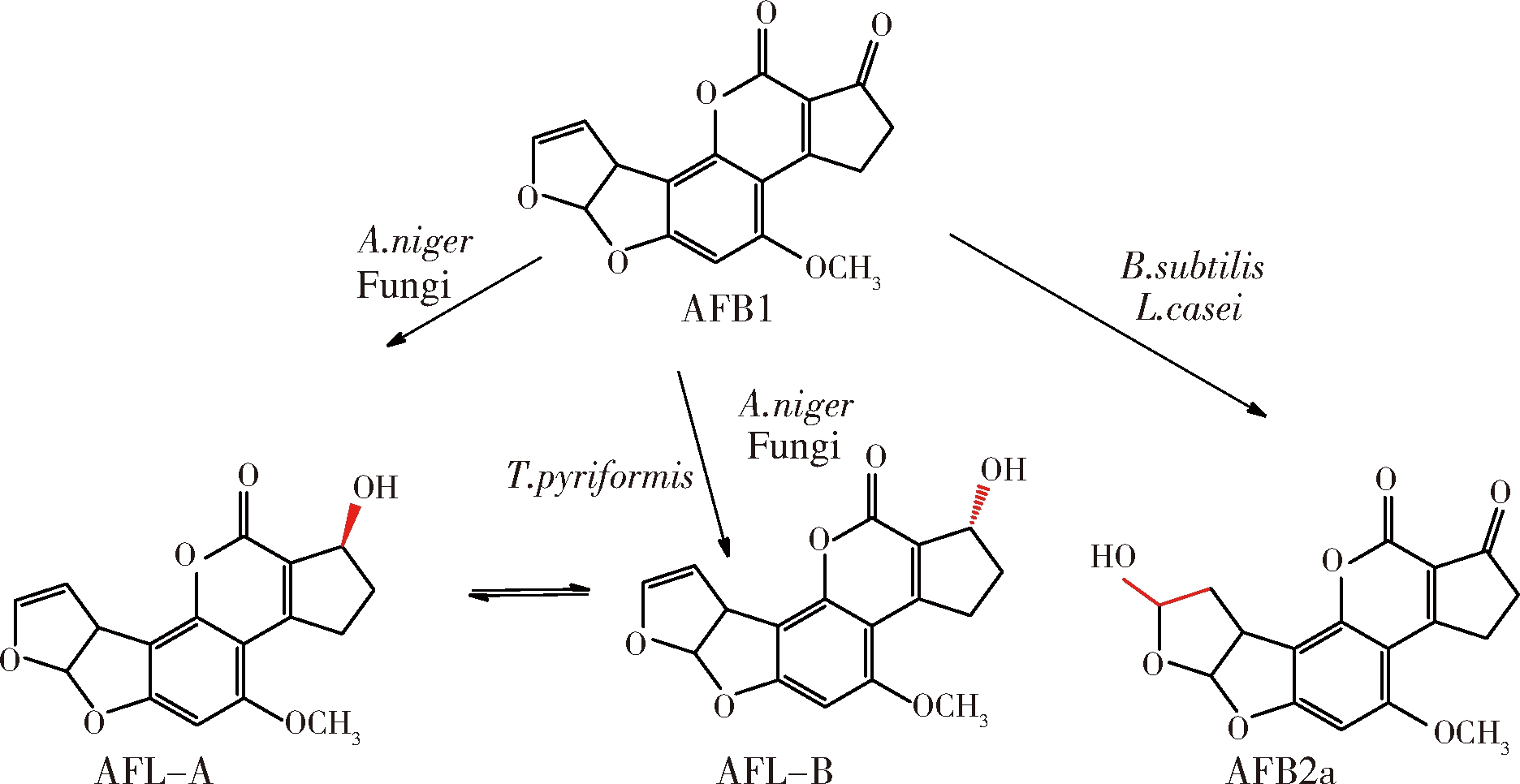
图2 部分真菌降解AFB1的产物
Fig.2 Degradation of AFB1 by certain fungi
此外,多种微生物可以降解ZEA。从瘤胃中分离的1株假单胞菌THN1可以将ZEA(2 μg/mL)作为唯一碳源完全降解利用[32];芽孢杆菌在LB培养基中发酵3 d后可以完全降解浓度为7.45 μg/mL的ZEA[33],且后续报道的地衣芽孢杆菌和短小芽孢杆菌对ZEA的降解率均高于85%[34-35](表2);而乳酸菌降解ZEA(1 μg/mL)最高效率仅为47.8%且过程较为缓慢,达到45%的降解率需要48 h以上[36]。一些研究推测了降解ZEA的途径,如酵母和假丝酵母可以将之还原为玉米赤霉烯醇(zearalenol, ZEL)[37];丝孢酵母对ZEA进行脱羧处理,使之不再具有雌激素类似结构从而消除毒性(图3)[38]。纳豆芽孢杆菌降解ZEA的途径如图3所示,先将ZEA中羰基位置氧化成内酯环,然后通过水解改变其雌激素类似结构,达到脱毒效果[39]。ZEA也可以通过糖基化、磺酸化等方式降低毒性,但是根据动物实验显示,这些结构在肠道中会重新水解还原成ZEA,因此不能够认为是有效的脱毒手段[40]。
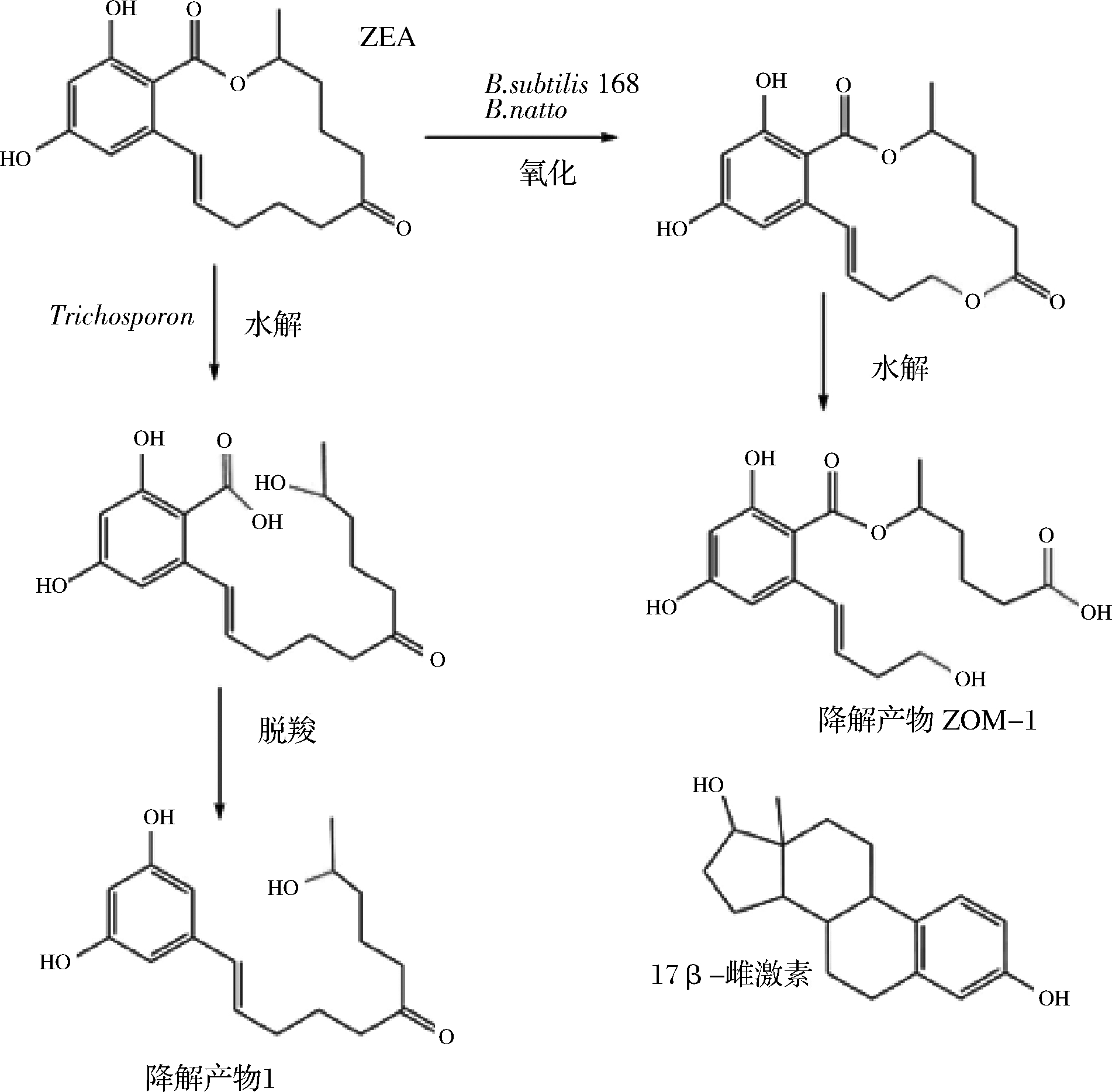
图3 部分微生物对 ZEA的降解途径
Fig.3 Degradation of ZEA by some microorganisms
伏马毒素是一类具有长链氨基酸骨架的二酯,它的降解是经过羧酸酯酶和氨基转移酶等多步水解完成的[13]。黑色酵母、棘状霉菌等都可以通过水解酯键降解伏马毒素B1(FB1)[41]。最近发现,从蘑菇堆肥中分离出的混合菌种,其中假单胞菌占主要成分,可以对培养基中伏马毒素B1(10 μg/mL)进行完全降解,采用液相色谱飞行时间质谱对代谢产物进行鉴定,并通过细胞实验证明了降解产物是一种低毒的物质[42](见图4),通过羧酸酯酶脱去2个分子的己三酸生成部分水解伏马毒素B1(hydrolyzed fumonisin B1, HFB1),再通过转氨酶作用脱去氨基,形成低毒性的降解产物2-酮-HFB1。
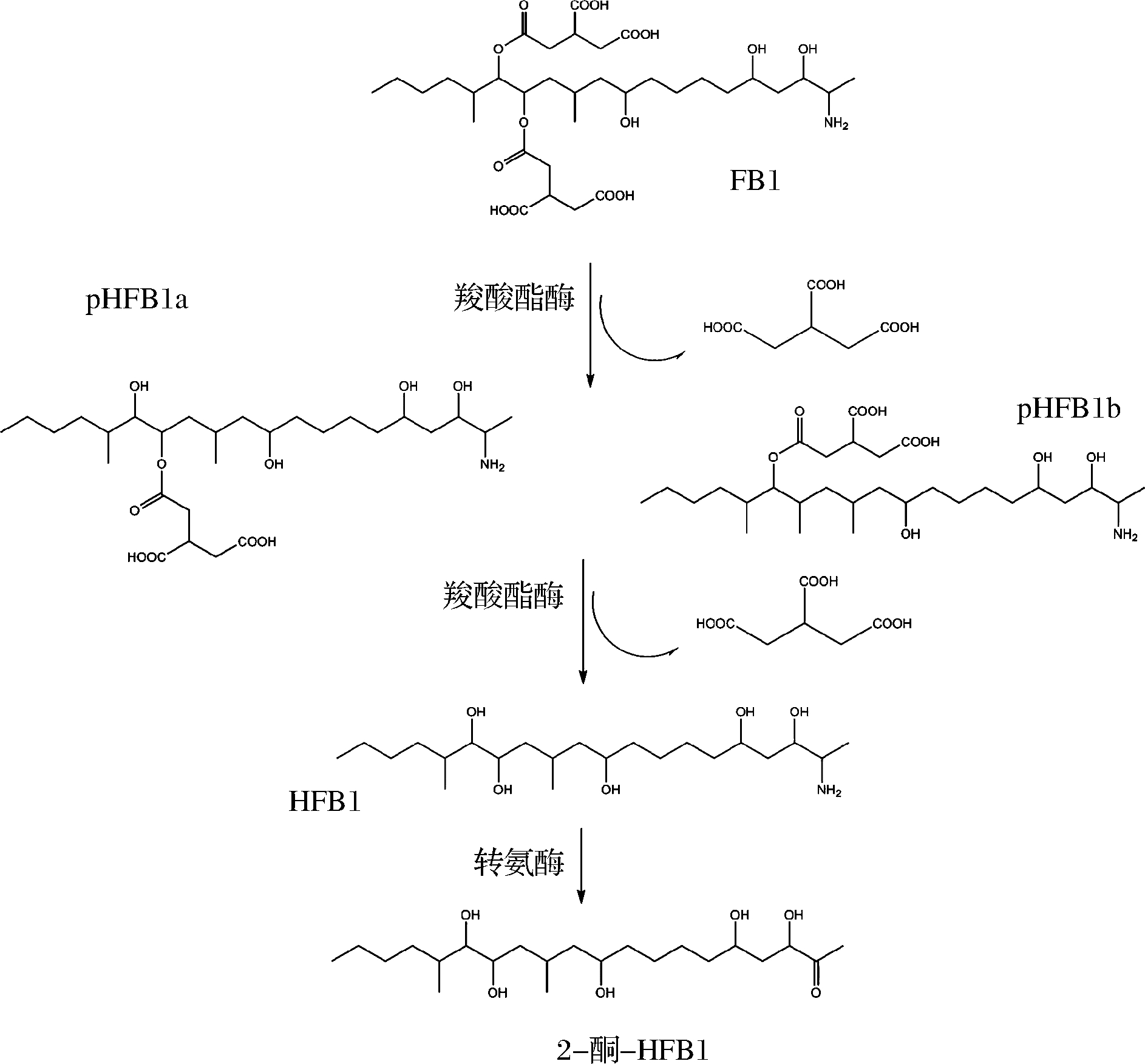
图4 假单胞菌降解伏马毒素B1的途径
Fig.4 Degradation diagram for fumonisin B1 by Pseudomona
根霉和毛霉等真菌能够将液体培养基中的OTA降解到一个极低的水平,红球菌和短杆菌等也能够降解90%以上的OTA,但需要5~10 d [43-44]。从动物肠道分离出一些厌氧微生物可以快速降解OTA,在39 ℃条件下24 h内即可降解75%以上的OTA(0.1~1 μg/mL),尤其是真细菌属微生物可以在6 h内降解95%的OTA(0.2 μg/mL)[45]。有学者证明水解OTA中的内酯环可达到降低毒性的效果[24](图5)。

图5 微生物对OTA的两种主要降解途径
Fig.5 Two main pathways of OTA degradation by microorganisms
从贮藏玉米中分离出1株芽孢杆菌可以通过另一种方式降解OTA,经实验证明其分泌的羧肽酶对OTA中的肽键进行了水解,生成低毒的代谢产物OT-α(图5)[46]。此外,小片球菌、黑曲霉、不动杆菌等分泌的羧肽酶A、蛋白酶A、脂肪酶和OTA酶等胞外酶和酵母分泌的胞内酶都通过此途径降解OTA,且被认为是最有效的降解途径[47](表2)。
单端孢霉烯类毒素的主要毒性结构在于环氧结构和特殊位置的羟基或乙酰基,因此,去环氧化、去乙酰化或者糖基化是微生物脱毒方式[48]。GAO等从鸡肠道中分离出1株细菌,以脱氧雪腐烯醇为底物时可以氧化脱除其分子上的环氧结构,对于T-2毒素、HT-2 和T-2三醇等都具有相似的作用方式[49]。土壤中分离得到的包含梭菌属、柠檬酸杆菌属、肠球菌属、链霉菌属和单胞菌属的混合菌种在12~40 ℃、pH 6.0~7.5的条件下可以使DON完全去环氧化[50]。HE等也做了相似的研究,从土壤中分离出的假单胞菌在48 h内完全降解DON(100 μg/mL)生成3-去环氧-DON,同样也可以将DON或雪腐烯醇(nivalenol, NIV)变为3-酮-DON或3-酮-NIV[51](图6);乳酸菌分泌胞外酶通过去环氧化对DON和T-2毒素进行降解,降解率分别为100%和58%[9, 52](表2)。
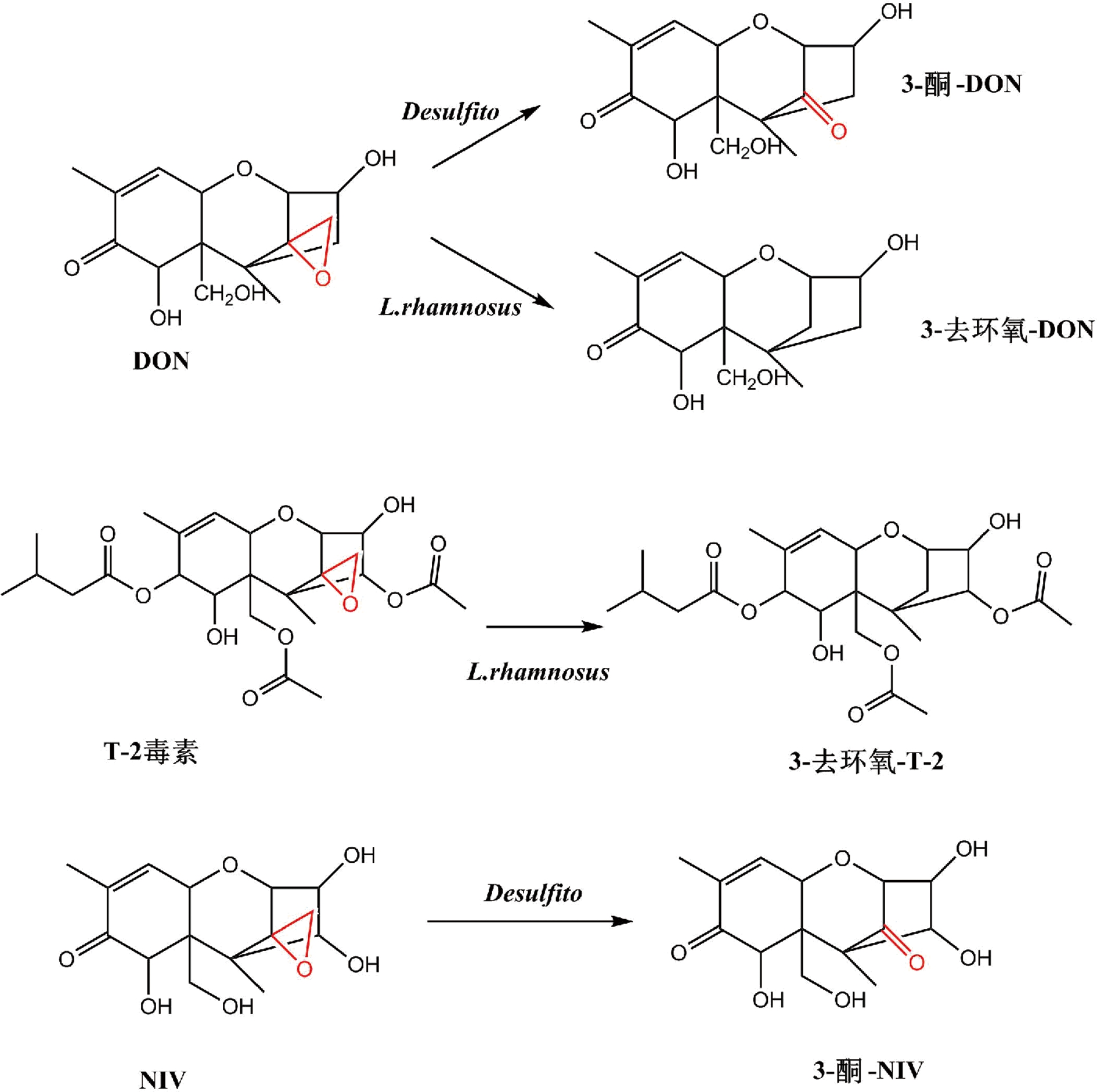
图6 单端孢霉烯类毒素的主要降解途径
Fig.6 Major pathways of trichothecene degradation
由于细菌可分泌多种酶,一些细菌可以同时降解多种毒素。如PETCHKONGKAEW等[53]从发酵黄豆中分离出1株芽孢杆菌可以降解AFB1和OTA。短杆菌属中有多种细菌可以降解多酚芳环结构,并分泌羧肽酶可以用于降解AFs和OTs[43];红球菌可以同时降解AFB1、ZEA和OTA等,与其分泌的内酯水解酶有关[27]。
2 研究展望
生物吸附通过微生物细胞壁上的多糖和蛋白成分对霉菌毒素进行结合,被吸附后的毒素即使经过胃肠道的消化也不易被释放,但目前针对霉菌毒素释放的实验多数为体外模拟,缺乏相应的动物实验进行效果评价。
生物降解是未来的研究热点,许多菌种被证明能降解霉菌毒素,但机理与途径并不清楚,需要进一步探究降解方式、降解产物毒性以及系统的评价方法。另外,生物降解霉菌毒素周期大多在12~60 h,有些发酵降解甚至长达30 d,而降解酶分离纯化较为困难,大多数稳定性较差,难以满足生产过程中快速高效的需求,还需进一步研究其他快速高效降解的方法。
目前的研究主要集中在单株菌降解单一种类霉菌毒素,而农作物污染的霉菌毒素往往不止1种且它们之间具有协同增强毒性的作用,因此对单一毒素的脱除研究并不能很好地满足实际需求,需要进一步加强对多种霉菌毒素同时进行降解的研究。理论上,生物降解毒素需要一整套完整的酶系才能彻底解毒,通过多菌种的合理复配,优化发酵条件,使分泌的各类酶协同增效、彻底降解同种或多种霉菌毒素也是未来生物降解的发展趋势。
[1] HATHOUT A S, ALY S E.Biological detoxification of mycotoxins: a review[J].Annals of Microbiology, 2014, 64(3): 905-919.
[2] CAO H, ZHI Y, XU H, et al.Zearalenone causes embryotoxicity and induces oxidative stress and apoptosis in differentiated human embryonic stem cells[J].Toxicol In Vitro, 2019, 54:243-250.
[3] SCHULZ M C, SCHUMANN L, ROTTKORD U, et al.Synergistic action of the nephrotoxic mycotoxins ochratoxin A and citrinin at nanomolar concentrations in human proximal tubule-derived cells[J].Toxicol Lett, 2018, 291:149-157.
[4] LUO Y, LIU X, LI J.Updating techniques on controlling mycotoxins-A review[J].Food Control, 2018, 89:123-132.
[5] GAVAHIAN M, CULLEN P J.Cold plasma as an emerging technique for mycotoxin-free food: Efficacy, mechanisms, and trends[J].Food Reviews International, 2019,1: 1-22.
[6] PANKAJ S K, SHI H, KEENER K M.A review of novel physical and chemical decontamination technologies for aflatoxin in food[J].Trends in Food Science & Technology, 2018, 71:73-83.
[7] ALSHANNAQ A, YU J H.Occurrence, toxicity, and analysis of major mycotoxins in food[J].Int J Environ Res Public Health, 2017, 14(6):632-651.
[8] HASKARD C A, EL-NEZAMI H S, KANKAANPAA P E, et al.Surface binding of aflatoxin B(1)by lactic acid bacteria[J].Appl Environ Microbiol, 2001, 67(7): 3 086-3 091.
[9] JUODEIKIENE G, BARTKIENE E, CERNAUSKAS D, et al.Antifungal activity of lactic acid bacteria and their application for Fusarium mycotoxin reduction in malting wheat grains[J].Lwt, 2018, 89:307-314.
[10] NIDERKORN V, MORGAVI D P, ABOAB B, et al.Cell wall component and mycotoxin moieties involved in the binding of fumonisin B1 and B2 by lactic acid bacteria[J].J Appl Microbiol, 2009, 106(3): 977-985.
[11] KUHARIC Z, JAKOPOVIC Z, CANAK I, et al.Removing aflatoxin M1 from milk with native lactic acid bacteria, centrifugation, and filtration[J].Arh Hig Rada Toksikol, 2018, 69(4): 334-339.
[12] MARTINEZ M P, MAGNOLI A P, GONZALEZ PEREYRA M L, et al.Probiotic bacteria and yeasts adsorb aflatoxin M1 in milk and degrade it to less toxic AFM1-metabolites[J].Toxicon, 2019,172:1-7.
[13] CHLEBICZ A, SLIZEWSKA K.In vitro detoxification of aflatoxin B1, deoxynivalenol, fumonisins, T-2 toxin and zearalenone by probiotic bacteria from genus Lactobacillus and Saccharomyces cerevisiae yeast[J].Probiotics Antimicrob Proteins, 2019.
[14] TAHEUR F B, FEDHILA K, CHAIEB K, et al.Adsorption of aflatoxin B1, zearalenone and ochratoxin A by microorganisms isolated from Kefir grains[J].Int J Food Microbiol, 2017, 251:1-7.
[15] ZHAI Y, HU S, ZHONG L, et al.Characterization of deoxynivalenol detoxification by Lactobacillus paracasei LHZ-1 isolated from yogurt[J].J Food Prot, 2019, 82(8): 1 292-1 299.
[16] BEJAOUI H, MATHIEU F, TAILLANDIER P, et al.Ochratoxin A removal in synthetic and natural grape juices by selected oenological Saccharomyces strains[J].J Appl Microbiol, 2004, 97(5): 1 038-1 044.
[17] ROGOWSKA A, POMASTOWSKI P, WALCZAK J, et al.Investigation of zearalenone adsorption and biotransformation by microorganisms cultured under cellular stress conditions[J].Toxins(Basel), 2019, 11(8):463-480.
[18] LIEW W P, NURUL-ADILAH Z, THAN L T L, et al.The binding efficiency and interaction of Lactobacillus casei Shirota toward aflatoxin B1[J].Front Microbiol, 2018, 9:1 503-1 514.
[19] ARMANDO M R, PIZZOLITTO R P, DOGI C A, et al.Adsorption of ochratoxin A and zearalenone by potential probiotic Saccharomyces cerevisiae strains and its relation with cell wall thickness[J].J Appl Microbiol, 2012, 113(2): 256-264.
[20] DAWLAL P, BRABET C, THANTSHA M S, et al.Visualisation and quantification of fumonisins bound by lactic acid bacteria isolates from traditional African maize-based fermented cereals, ogi and mahewu[J].Food Addit Contam Part A Chem Anal Control Expo Risk Assess, 2019, 36(2): 296-307.
[21] AAZAMI M H, NASRI M H F, MOJTAHEDI M, et al.In vitro aflatoxin B1 binding by the cell wall and(1-->3)-beta-d-Glucan of Baker's Yeast[J].J Food Prot, 2018, 81(4): 670-676.
[22] SEVIM S, TOPAL G G, TENGILIMOGLU-METIN M M, et al.Effects of inulin and lactic acid bacteria strains on aflatoxin M1 detoxification in yoghurt[J].Food Control, 2019, 100:235-239.
[23] KROL A, POMASTOWSKI P, RAFINSKA K, et al.Microbiology neutralization of zearalenone using Lactococcus lactis and Bifidobacterium sp.[J].Anal Bioanal Chem, 2018, 410(3): 943-952.
[24] SHI L, LIANG Z, LI J, et al.Ochratoxin A biocontrol and biodegradation by Bacillus subtilis CW 14[J].J Sci Food Agric, 2014, 94(9): 1 879-1 885.
[25] RUIYU Z.Inhibiting Aspergillus flavus growth and degrading aflatoxin B1 by combined beneficial microbes[J].African Journal of Biotechnology, 2012, 11(65): 12 903-12 909.
[26] GAO X, MA Q, ZHAO L, et al.Isolation of Bacillus subtilis: screening for aflatoxins B1, M1, and G1 detoxification[J].European Food Research and Technology, 2011, 232(6): 957-962.
[27] RISA A, DIVINYI D M, BAKA E, et al.Aflatoxin B1 detoxification by cell-free extracts of Rhodococcus strains[J].Acta Microbiol Immunol Hung, 2017, 64(4): 423-438.
[28] MWAKINYALI S E, MING Z, XIE H, et al.Investigation and characterization of Myroides odoratimimus strain 3J2MO aflatoxin B1 degradation[J].J Agric Food Chem, 2019, 67(16): 4 595-4 602.
[29] LIU D L, YAO D S, LIANG R, et al.Detoxification of aflatoxin B1 by enzymes isolated from Armillariella tabescens[J].Food Chem Toxicol, 1998, 36(7): 563-574.
[30] ALBERTS J F, GELDERBLOM W C, BOTHA A, et al.Degradation of aflatoxin B(1)by fungal laccase enzymes[J].Int J Food Microbiol, 2009, 135(1): 47-52.
[31] NAKAZATO M, MOROZUMI S, SAITO K, et al.Interconversion of aflatoxin B1 and aflatoxicol by several fungi[J].Appl Environ Microbiol, 1990, 56(5): 1 465-1 470.
[32] TAN H, ZHANG Z, HU Y, et al.Isolation and characterization of Pseudomonas otitidis TH-N1 capable of degrading Zearalenone[J].Food Control, 2015, 47:285-290.
[33] WANG N, LI P, PAN J, et al.Bacillus velezensis A2 fermentation exerts a protective effect on renal injury induced by Zearalenone in mice[J].Sci Rep, 2018, 8(1): 13 646-13 659.
[34] WANG G, YU M, DONG F, et al.Esterase activity inspired selection and characterization of zearalenone degrading bacteria Bacillus pumilus ES-21[J].Food Control, 2017, 77:57-64.
[35] YI P J, PAI C K, LIU J R.Isolation and characterization of a Bacillus licheniformis strain capable of degrading zearalenone[J].World Journal of Microbiology and Biotechnology, 2010, 27(5): 1 035-1 043.
[36] ZHAO L, JIN H, LAN J, et al.Detoxification of zearalenone by three strains of Lactobacillus plantarum from fermented food in vitro[J].Food Control, 2015, 54:158-164.
[37] WANG N, WU W, PAN J, et al.Detoxification strategies for zearalenone using microorganisms: A review[J].Microorganisms, 2019, 7(7): 208-221.
[38] VEKIRU E, HAMETNER C, MITTERBAUER R, et al.Cleavage of zearalenone by Trichosporon mycotoxinivorans to a novel nonestrogenic metabolite[J].Appl Environ Microbiol, 2010, 76(7): 2 353-2 359.
[39] TINYIRO S E, WOKADALA C, XU D, et al.Adsorption and degradation of zearalenone by Bacillus strains[J].Folia Microbiol(Praha), 2011, 56(4): 321-327.
[40] BEN SALEM I, BOUSSABBEH M, PIRES DA SILVA J, et al.SIRT1 protects cardiac cells against apoptosis induced by zearalenone or its metabolites alpha- and beta-zearalenol through an autophagy-dependent pathway[J].Toxicol Appl Pharmacol, 2017, 314:82-90.
[41] HARTINGER D, MOLL W.Fumonisin elimination and prospects for detoxification by enzymatic transformation[J].World Mycotoxin Journal, 2011, 4(3): 271-283.
[42] ZHAO Z, ZHANG Y, GONG A, et al.Biodegradation of mycotoxin fumonisin B1 by a novel bacterial consortium SAAS79[J].Appl Microbiol Biotechnol, 2019, 103(17): 7 129-7 140.
[43] RODRIGUEZ H, REVERON I, DORIA F, et al.Degradation of ochratoxin a by Brevibacterium species[J].J Agric Food Chem, 2011, 59(19): 10 755-10 760.
[44] VARGA J, PETERI Z, TABORI K, et al.Degradation of ochratoxin A and other mycotoxins by Rhizopus isolates[J].Int J Food Microbiol, 2005, 99(3): 321-328.
[45] CHEN W, LI C, ZHANG B, et al.Advances in biodetoxification of ochratoxin A-A review of the past five decades[J].Front Microbiol, 2018, 9:1 386.
[46] CHANG X, WU Z, WU S, et al.Degradation of ochratoxin A by Bacillus amyloliquefaciens ASAG1[J].Food Addit Contam Part A Chem Anal Control Expo Risk Assess, 2015, 32(4): 564-571.
[47] ZHANG X, YANG H, APALIYA M T, et al.The mechanisms involved in ochratoxin A elimination by Yarrowia lipolytica Y-2[J].Annals of Applied Biology, 2018, 173(2): 164-174.
[48] LOI M, FANELLI F, LIUZZI V C, et al.Mycotoxin biotransformation by native and commercial enzymes: Present and future perspectives[J].Toxins(Basel), 2017, 9(4):111.
[49] GAO X, MU P, WEN J, et al.Detoxification of trichothecene mycotoxins by a novel bacterium, Eggerthella sp.DII-9[J].Food Chem Toxicol, 2018, 112:310-319.
[50] ISLAM R, ZHOU T, YOUNG J C, et al.Aerobic and anaerobic de-epoxydation of mycotoxin deoxynivalenol by bacteria originating from agricultural soil[J].World J Microbiol Biotechnol, 2012, 28(1): 7-13.
[51] HE W J, YUAN Q S, ZHANG Y B, et al.Aerobic de-epoxydation of trichothecene mycotoxins by a soil bacterial consortium isolated using in situ soil enrichment[J].Toxins(Basel), 2016, 8(10): 277-289.
[52] QU R, JIANG C, WU W, et al.Conversion of DON to 3-epi-DON in vitro and toxicity reduction of DON in vivo by Lactobacillus rhamnosus[J].Food Funct, 2019, 10(5): 2 785-2 796.
[53] PETCHKONGKAEW A, TAILLANDIER P, GASALUCK P, et al.Isolation of Bacillus spp.from Thai fermented soybean(Thua-nao): screening for aflatoxin B1 and ochratoxin A detoxification[J].J Appl Microbiol, 2008, 104(5): 1 495-1 502.